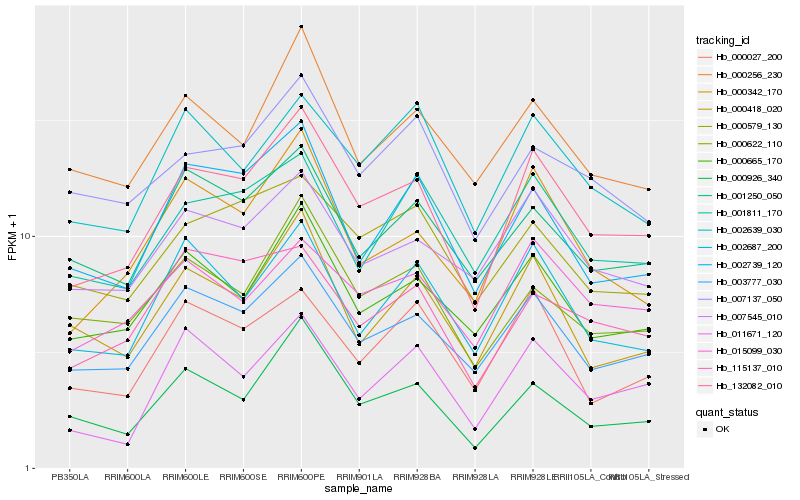
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_132082_010 |
0.0 |
- |
- |
hypothetical protein VITISV_030895 [Vitis vinifera] |
| 2 |
Hb_003777_030 |
0.0720695649 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 3 |
Hb_000342_170 |
0.0885037697 |
- |
- |
PREDICTED: uncharacterized protein LOC100807540 isoform X2 [Glycine max] |
| 4 |
Hb_000665_170 |
0.0893830455 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 5 |
Hb_007545_010 |
0.0963157051 |
- |
- |
HIV-1 rev binding protein, hrbl, putative [Ricinus communis] |
| 6 |
Hb_002739_120 |
0.1021559998 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 7 |
Hb_000926_340 |
0.1025868903 |
- |
- |
PREDICTED: EH domain-containing protein 1 isoform X1 [Jatropha curcas] |
| 8 |
Hb_002687_200 |
0.1028021853 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_015099_030 |
0.1030348731 |
- |
- |
PREDICTED: uncharacterized protein LOC105649867 [Jatropha curcas] |
| 10 |
Hb_000256_230 |
0.1049514651 |
- |
- |
PREDICTED: uncharacterized protein LOC105637594 [Jatropha curcas] |
| 11 |
Hb_001250_050 |
0.1056878977 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 12 |
Hb_000418_020 |
0.1062230215 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 13 |
Hb_001811_170 |
0.1062738439 |
- |
- |
dynamin, putative [Ricinus communis] |
| 14 |
Hb_000579_130 |
0.1066749883 |
- |
- |
ubiquitin-protein ligase, putative [Ricinus communis] |
| 15 |
Hb_000622_110 |
0.1074640221 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 16 |
Hb_002639_030 |
0.1089053308 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 17 |
Hb_115137_010 |
0.1102104551 |
- |
- |
PREDICTED: probable protein S-acyltransferase 23 isoform X2 [Jatropha curcas] |
| 18 |
Hb_011671_120 |
0.1118037771 |
- |
- |
hypothetical protein JCGZ_26275 [Jatropha curcas] |
| 19 |
Hb_007137_050 |
0.1121643084 |
- |
- |
Uncharacterized protein isoform 1 [Theobroma cacao] |
| 20 |
Hb_000027_200 |
0.112777951 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |