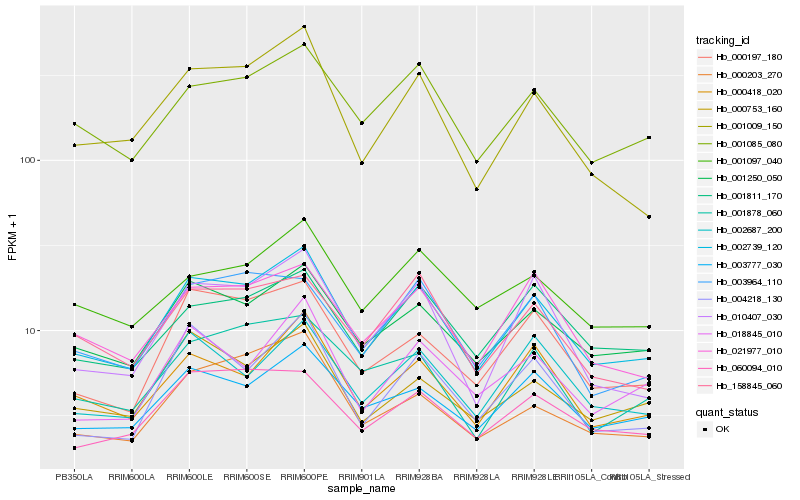
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002739_120 |
0.0 |
- |
- |
PREDICTED: zinc finger protein ZPR1-like [Jatropha curcas] |
| 2 |
Hb_001250_050 |
0.068485757 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 3 |
Hb_001085_080 |
0.0776074086 |
- |
- |
eukaryotic translation elongation factor 1-alpha [Hevea brasiliensis] |
| 4 |
Hb_003777_030 |
0.0814116514 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 5 |
Hb_000418_020 |
0.0817246816 |
- |
- |
PREDICTED: sulfhydryl oxidase 2 isoform X1 [Jatropha curcas] |
| 6 |
Hb_000197_180 |
0.0820561752 |
- |
- |
PREDICTED: bifunctional dihydrofolate reductase-thymidylate synthase 1 [Jatropha curcas] |
| 7 |
Hb_001009_150 |
0.082458819 |
- |
- |
Glyceraldehyde-3-phosphate dehydrogenase [Gossypium arboreum] |
| 8 |
Hb_021977_010 |
0.0850072844 |
- |
- |
PREDICTED: leukotriene A-4 hydrolase homolog [Jatropha curcas] |
| 9 |
Hb_010407_030 |
0.0851986203 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 10 |
Hb_002687_200 |
0.0919393114 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_001811_170 |
0.0925316889 |
- |
- |
dynamin, putative [Ricinus communis] |
| 12 |
Hb_000753_160 |
0.0928126856 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 13 |
Hb_158845_060 |
0.093153779 |
- |
- |
PREDICTED: nudix hydrolase 3 isoform X1 [Jatropha curcas] |
| 14 |
Hb_060094_010 |
0.0939736297 |
- |
- |
PREDICTED: C2 and GRAM domain-containing protein At1g03370 isoform X2 [Jatropha curcas] |
| 15 |
Hb_000203_270 |
0.094042461 |
- |
- |
alpha-xylosidase, putative [Ricinus communis] |
| 16 |
Hb_001878_060 |
0.0943908324 |
transcription factor |
TF Family: G2-like |
hypothetical protein POPTR_0007s15110g [Populus trichocarpa] |
| 17 |
Hb_018845_010 |
0.0952564921 |
- |
- |
PREDICTED: uncharacterized protein LOC105642055 [Jatropha curcas] |
| 18 |
Hb_001097_040 |
0.096118085 |
- |
- |
PREDICTED: probable dolichyl-diphosphooligosaccharide--protein glycosyltransferase subunit 3B [Jatropha curcas] |
| 19 |
Hb_004218_130 |
0.0973057049 |
- |
- |
PREDICTED: uridine 5'-monophosphate synthase [Jatropha curcas] |
| 20 |
Hb_003964_110 |
0.0973282827 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g26460, mitochondrial [Jatropha curcas] |