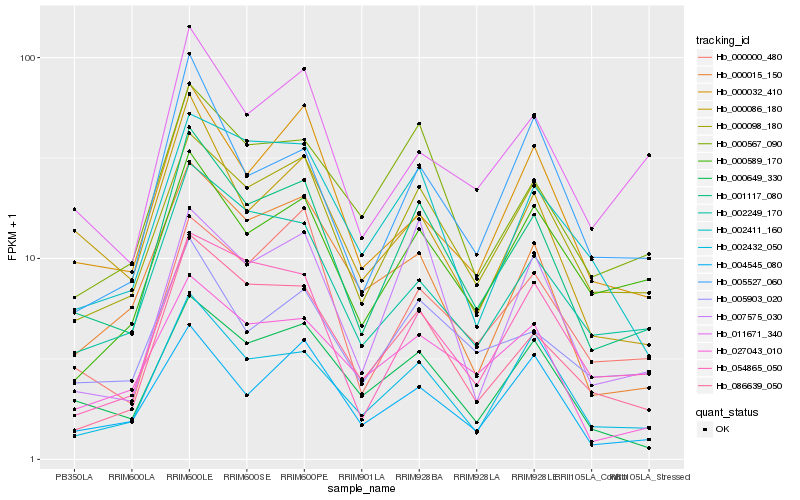
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001117_080 |
0.0 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase 2 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000098_180 |
0.0919278608 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 3 |
Hb_086639_050 |
0.1046302558 |
- |
- |
PREDICTED: uncharacterized protein At1g04910 isoform X1 [Jatropha curcas] |
| 4 |
Hb_002432_050 |
0.11134419 |
- |
- |
PREDICTED: uncharacterized protein LOC105648523 [Jatropha curcas] |
| 5 |
Hb_000649_330 |
0.1123085889 |
- |
- |
hypothetical protein POPTR_0010s11880g [Populus trichocarpa] |
| 6 |
Hb_000567_090 |
0.1157077209 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_002249_170 |
0.116370986 |
- |
- |
copine, putative [Ricinus communis] |
| 8 |
Hb_005903_020 |
0.1219054128 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 9 |
Hb_054865_050 |
0.1231331785 |
- |
- |
calcium-dependent protein kinase, putative [Ricinus communis] |
| 10 |
Hb_000086_180 |
0.1250190003 |
- |
- |
hypothetical protein JCGZ_17674 [Jatropha curcas] |
| 11 |
Hb_027043_010 |
0.1251808287 |
- |
- |
PREDICTED: glycerophosphodiester phosphodiesterase GDPD4 [Jatropha curcas] |
| 12 |
Hb_000032_410 |
0.12649056 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 13 |
Hb_011671_340 |
0.1287000203 |
- |
- |
small GTPase [Hevea brasiliensis] |
| 14 |
Hb_000015_150 |
0.1288817596 |
- |
- |
Ubiquitin-conjugating enzyme E2 7 family protein [Populus trichocarpa] |
| 15 |
Hb_000000_480 |
0.1291616553 |
- |
- |
PREDICTED: deSI-like protein At4g17486 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004545_080 |
0.1293530398 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_007575_030 |
0.1315298496 |
- |
- |
PREDICTED: protein RCC2 homolog [Jatropha curcas] |
| 18 |
Hb_005527_060 |
0.1316477723 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 19 |
Hb_002411_160 |
0.1325937627 |
- |
- |
PREDICTED: uncharacterized protein LOC105631262 isoform X1 [Jatropha curcas] |
| 20 |
Hb_000589_170 |
0.1330297293 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |