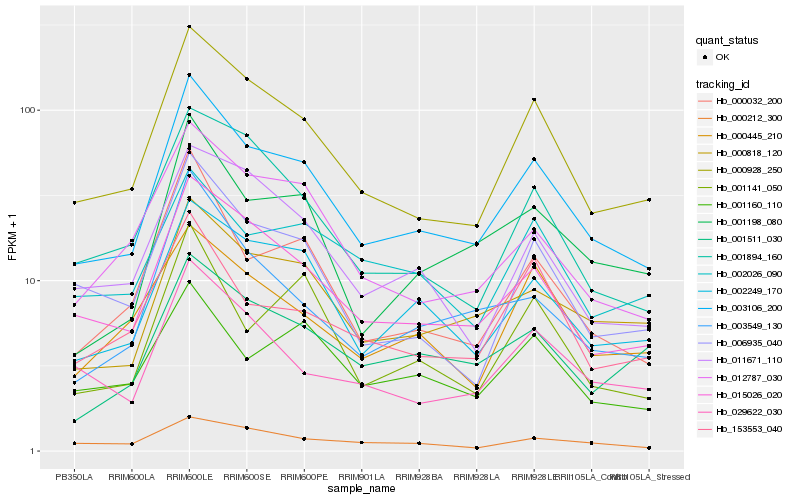
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_003106_200 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640430 isoform X1 [Jatropha curcas] |
| 2 |
Hb_000928_250 |
0.0803703474 |
- |
- |
calcium ion binding protein, putative [Ricinus communis] |
| 3 |
Hb_015026_020 |
0.0892673878 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing protein Os07g0563300 [Jatropha curcas] |
| 4 |
Hb_000818_120 |
0.0988633776 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_002249_170 |
0.1013647355 |
- |
- |
copine, putative [Ricinus communis] |
| 6 |
Hb_000032_200 |
0.1082188122 |
- |
- |
PREDICTED: probable polygalacturonase [Jatropha curcas] |
| 7 |
Hb_001198_080 |
0.1114660644 |
- |
- |
PREDICTED: uncharacterized protein LOC105633342 isoform X2 [Jatropha curcas] |
| 8 |
Hb_012787_030 |
0.1154388421 |
- |
- |
PREDICTED: dnaJ homolog subfamily B member 1 [Jatropha curcas] |
| 9 |
Hb_006935_040 |
0.1226018115 |
- |
- |
PREDICTED: uncharacterized protein LOC105629236 [Jatropha curcas] |
| 10 |
Hb_001894_160 |
0.1240189515 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_003549_130 |
0.1267027099 |
- |
- |
neutral/alkaline invertase [Manihot esculenta] |
| 12 |
Hb_029622_030 |
0.1300426761 |
- |
- |
ankyrin repeat-containing protein, putative [Ricinus communis] |
| 13 |
Hb_153553_040 |
0.1302158826 |
- |
- |
hypothetical protein JCGZ_19478 [Jatropha curcas] |
| 14 |
Hb_001160_110 |
0.1305233459 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 15 |
Hb_011671_110 |
0.1305261874 |
- |
- |
PREDICTED: uncharacterized protein LOC105648628 [Jatropha curcas] |
| 16 |
Hb_001141_050 |
0.132822459 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 17 |
Hb_001511_030 |
0.1362591556 |
- |
- |
PREDICTED: inactive receptor-like serine/threonine-protein kinase At2g40270 [Jatropha curcas] |
| 18 |
Hb_000212_300 |
0.1363128564 |
- |
- |
PREDICTED: thaumatin-like protein [Jatropha curcas] |
| 19 |
Hb_000445_210 |
0.1375539382 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105639010 [Jatropha curcas] |
| 20 |
Hb_002026_090 |
0.1406224427 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2 4-like [Jatropha curcas] |