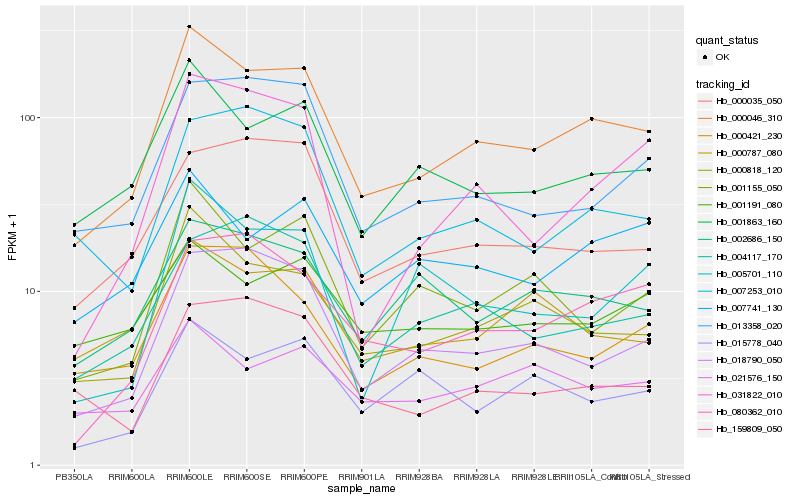
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000046_310 |
0.0 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 2 |
Hb_000818_120 |
0.1018706796 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_001863_160 |
0.113414562 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 4 |
Hb_018790_050 |
0.1318580774 |
- |
- |
hypothetical protein CICLE_v10017858mg [Citrus clementina] |
| 5 |
Hb_001155_050 |
0.1329674149 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 6 |
Hb_080362_010 |
0.1383730073 |
- |
- |
hypothetical protein JCGZ_14167 [Jatropha curcas] |
| 7 |
Hb_007741_130 |
0.1400362051 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 8 |
Hb_013358_020 |
0.1426088317 |
- |
- |
ccr4-associated factor, putative [Ricinus communis] |
| 9 |
Hb_002686_150 |
0.1431474776 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 10 |
Hb_000035_050 |
0.1461230851 |
- |
- |
PREDICTED: probable CCR4-associated factor 1 homolog 7 [Jatropha curcas] |
| 11 |
Hb_000787_080 |
0.1465333227 |
- |
- |
PREDICTED: TBC1 domain family member 16-like [Jatropha curcas] |
| 12 |
Hb_031822_010 |
0.1481664749 |
- |
- |
PREDICTED: uncharacterized protein LOC103496015 [Cucumis melo] |
| 13 |
Hb_000421_230 |
0.1489514069 |
- |
- |
retinal degeneration B beta, putative [Ricinus communis] |
| 14 |
Hb_004117_170 |
0.1494748616 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 15 |
Hb_007253_010 |
0.1499302482 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 16 |
Hb_021576_150 |
0.1504832021 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 17 |
Hb_001191_080 |
0.1519042791 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_015778_040 |
0.1522817685 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 19 |
Hb_005701_110 |
0.1532911499 |
- |
- |
PREDICTED: probable 1-acylglycerol-3-phosphate O-acyltransferase [Jatropha curcas] |
| 20 |
Hb_159809_050 |
0.1540002887 |
- |
- |
AT14A, putative [Ricinus communis] |