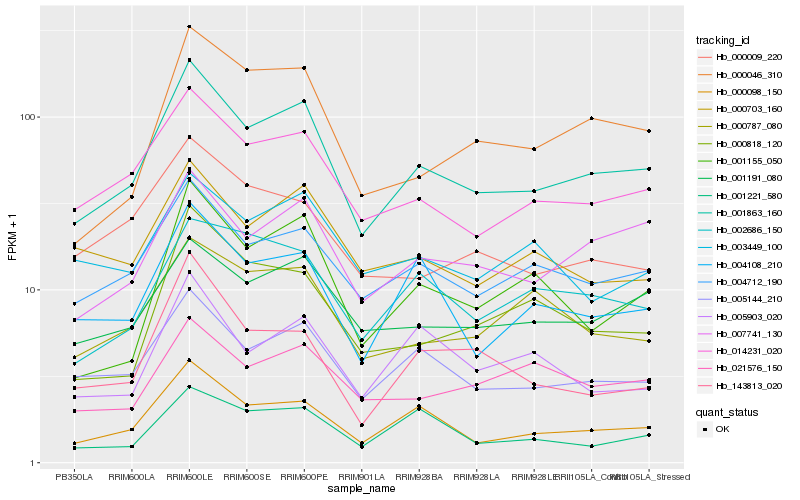
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001863_160 |
0.0 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 2 |
Hb_000098_150 |
0.0881617859 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 3 |
Hb_014231_020 |
0.0893824029 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 4 |
Hb_005144_210 |
0.0933052345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_004712_190 |
0.0941760007 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 6 |
Hb_007741_130 |
0.1034765548 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 7 |
Hb_001191_080 |
0.1100637827 |
- |
- |
protein with unknown function [Ricinus communis] |
| 8 |
Hb_000046_310 |
0.113414562 |
- |
- |
PREDICTED: inactive beta-amylase 9 [Jatropha curcas] |
| 9 |
Hb_004108_210 |
0.1153366892 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 10 |
Hb_001155_050 |
0.121210587 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 11 |
Hb_000703_160 |
0.1246786682 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_000787_080 |
0.1253170267 |
- |
- |
PREDICTED: TBC1 domain family member 16-like [Jatropha curcas] |
| 13 |
Hb_002686_150 |
0.1261804984 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 14 |
Hb_000818_120 |
0.1327659025 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000009_220 |
0.1328783171 |
- |
- |
PREDICTED: calcium uniporter protein 2, mitochondrial-like [Jatropha curcas] |
| 16 |
Hb_003449_100 |
0.1349304899 |
- |
- |
PREDICTED: protein YIPF6 homolog [Jatropha curcas] |
| 17 |
Hb_001221_580 |
0.1353361181 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 18 |
Hb_021576_150 |
0.1357026894 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 19 |
Hb_143813_020 |
0.1364715519 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 20 |
Hb_005903_020 |
0.1375322234 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |