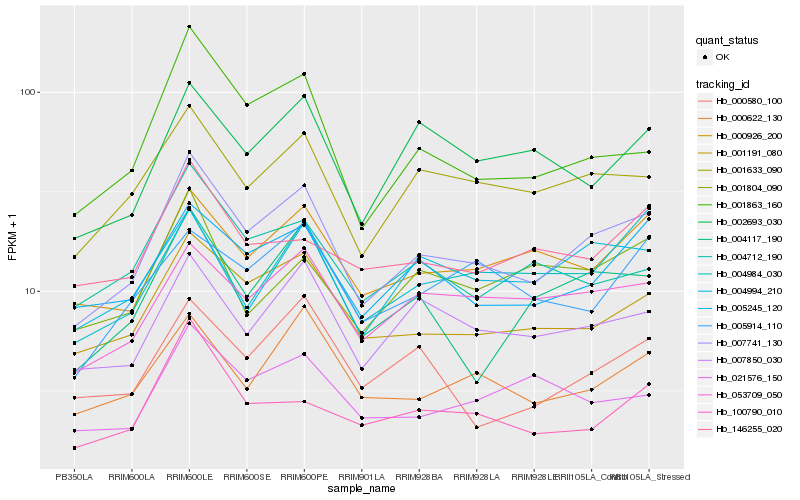
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007741_130 |
0.0 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 2 |
Hb_001191_080 |
0.0868308444 |
- |
- |
protein with unknown function [Ricinus communis] |
| 3 |
Hb_001633_090 |
0.0963132956 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 4 |
Hb_007850_030 |
0.1019767026 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 5 |
Hb_005245_120 |
0.10205375 |
- |
- |
XPA-binding protein, putative [Ricinus communis] |
| 6 |
Hb_001863_160 |
0.1034765548 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 7 |
Hb_053709_050 |
0.1056243227 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |
| 8 |
Hb_000926_200 |
0.1073222481 |
transcription factor |
TF Family: Rcd1-like |
PREDICTED: cell differentiation protein RCD1 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_004117_190 |
0.1108019892 |
- |
- |
PREDICTED: uncharacterized protein LOC105649109 [Jatropha curcas] |
| 10 |
Hb_000622_130 |
0.1109581533 |
- |
- |
PREDICTED: tubulin gamma-1 chain [Vitis vinifera] |
| 11 |
Hb_004712_190 |
0.119507367 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 12 |
Hb_002693_030 |
0.1214654749 |
- |
- |
PREDICTED: mitochondrial outer membrane protein porin of 34 kDa [Jatropha curcas] |
| 13 |
Hb_004994_210 |
0.1216979725 |
- |
- |
phosphomannomutase family protein [Populus trichocarpa] |
| 14 |
Hb_021576_150 |
0.1249511883 |
- |
- |
PREDICTED: formin-like protein 18 [Jatropha curcas] |
| 15 |
Hb_000580_100 |
0.125078925 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 16 |
Hb_004984_030 |
0.1291896049 |
- |
- |
protein with unknown function [Ricinus communis] |
| 17 |
Hb_146255_020 |
0.1301638699 |
- |
- |
PREDICTED: chromatin structure-remodeling complex protein BSH [Jatropha curcas] |
| 18 |
Hb_001804_090 |
0.1303210047 |
- |
- |
PREDICTED: uracil phosphoribosyltransferase isoform X1 [Jatropha curcas] |
| 19 |
Hb_005914_110 |
0.1309639646 |
- |
- |
PREDICTED: uncharacterized protein LOC105632180 [Jatropha curcas] |
| 20 |
Hb_100790_010 |
0.1312670178 |
- |
- |
conserved hypothetical protein [Ricinus communis] |