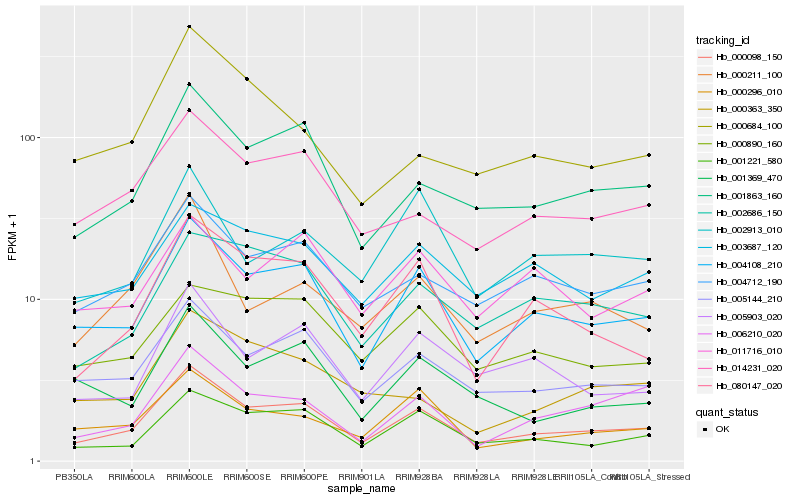
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000098_150 |
0.0 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 2 |
Hb_004108_210 |
0.0632929541 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 3 |
Hb_001863_160 |
0.0881617859 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 4 |
Hb_005144_210 |
0.092358907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001221_580 |
0.0965867614 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 6 |
Hb_004712_190 |
0.1056598256 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 7 |
Hb_014231_020 |
0.1145232749 |
- |
- |
PREDICTED: ras-related protein Rab11D [Jatropha curcas] |
| 8 |
Hb_080147_020 |
0.1155119367 |
- |
- |
3-hydroxyacyl-CoA dehyrogenase, putative [Ricinus communis] |
| 9 |
Hb_000296_010 |
0.1198955217 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 10 |
Hb_005903_020 |
0.1207875569 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 11 |
Hb_002686_150 |
0.125392148 |
- |
- |
Glucan endo-1,3-beta-glucosidase precursor, putative [Ricinus communis] |
| 12 |
Hb_003687_120 |
0.1291445134 |
- |
- |
PREDICTED: serine/threonine-protein kinase tricorner-like [Jatropha curcas] |
| 13 |
Hb_006210_020 |
0.1309867072 |
- |
- |
PREDICTED: calcium sensing receptor, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001369_470 |
0.1310147851 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 15 |
Hb_000211_100 |
0.1310252315 |
- |
- |
PREDICTED: biotin synthase [Jatropha curcas] |
| 16 |
Hb_000890_160 |
0.133406263 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 17 |
Hb_002913_010 |
0.139452147 |
- |
- |
PREDICTED: mitochondrial amidoxime reducing component 2-like isoform X1 [Jatropha curcas] |
| 18 |
Hb_011716_010 |
0.1422138159 |
- |
- |
PREDICTED: probable protein arginine N-methyltransferase 1.2 [Jatropha curcas] |
| 19 |
Hb_000684_100 |
0.1430349091 |
- |
- |
PREDICTED: cell number regulator 8 [Jatropha curcas] |
| 20 |
Hb_000363_350 |
0.1438630306 |
- |
- |
PREDICTED: transcription initiation factor TFIID subunit 6-like [Fragaria vesca subsp. vesca] |