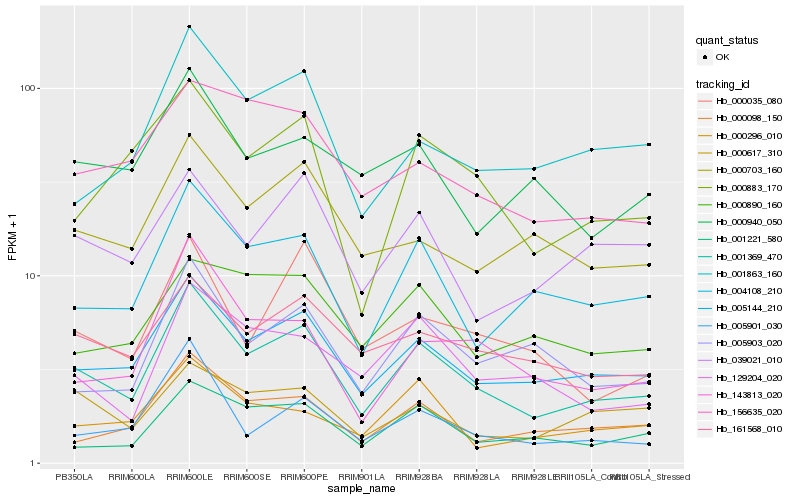
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_470 |
0.0 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 2 |
Hb_005144_210 |
0.0884725534 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 3 |
Hb_004108_210 |
0.1200574394 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 4 |
Hb_000098_150 |
0.1310147851 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 5 |
Hb_001221_580 |
0.1358867324 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 6 |
Hb_161568_010 |
0.1377556056 |
- |
- |
hydrolase, hydrolyzing O-glycosyl compounds, putative [Ricinus communis] |
| 7 |
Hb_005903_020 |
0.1383731597 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 8 |
Hb_000883_170 |
0.1390535257 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 9 |
Hb_143813_020 |
0.1400794395 |
transcription factor |
TF Family: WRKY |
hypothetical protein POPTR_0006s10950g [Populus trichocarpa] |
| 10 |
Hb_001863_160 |
0.140223287 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 11 |
Hb_000703_160 |
0.1405035357 |
- |
- |
PREDICTED: serine/threonine-protein kinase EDR1 isoform X2 [Jatropha curcas] |
| 12 |
Hb_129204_020 |
0.1432494129 |
- |
- |
PREDICTED: IAA-amino acid hydrolase ILR1-like 4 [Jatropha curcas] |
| 13 |
Hb_005901_030 |
0.147618192 |
- |
- |
PREDICTED: phragmoplast orienting kinesin 2 isoform X1 [Jatropha curcas] |
| 14 |
Hb_000617_310 |
0.1477921858 |
- |
- |
PREDICTED: cell cycle checkpoint control protein RAD9A [Jatropha curcas] |
| 15 |
Hb_039021_010 |
0.1483640591 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 16 |
Hb_156635_020 |
0.1504780187 |
- |
- |
PREDICTED: L-lactate dehydrogenase B [Jatropha curcas] |
| 17 |
Hb_000296_010 |
0.1530395508 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 18 |
Hb_000940_050 |
0.1536503323 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 19 |
Hb_000035_080 |
0.1558799808 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 20 |
Hb_000890_160 |
0.1585536452 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |