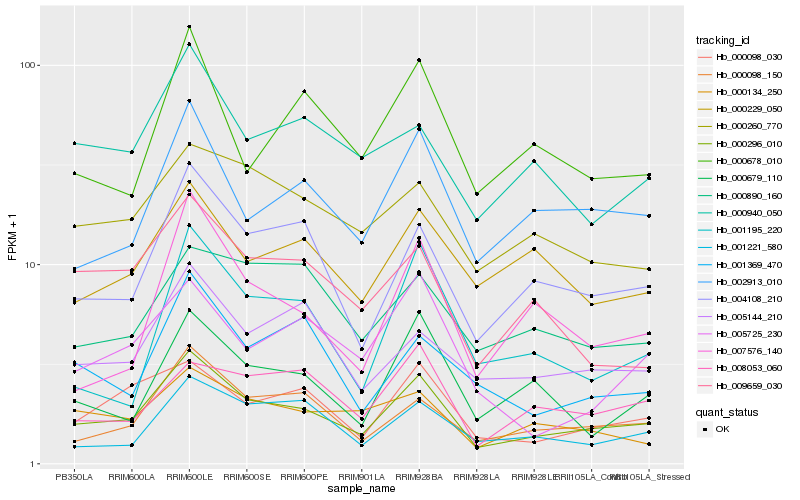
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000296_010 |
0.0 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 2 |
Hb_004108_210 |
0.1109648561 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 3 |
Hb_000098_150 |
0.1198955217 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 4 |
Hb_002913_010 |
0.1285700819 |
- |
- |
PREDICTED: mitochondrial amidoxime reducing component 2-like isoform X1 [Jatropha curcas] |
| 5 |
Hb_000098_030 |
0.1291968898 |
- |
- |
MLO-like protein 4 [Gossypium arboreum] |
| 6 |
Hb_005144_210 |
0.1410084668 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000940_050 |
0.1448544191 |
- |
- |
hypothetical protein JCGZ_19588 [Jatropha curcas] |
| 8 |
Hb_001221_580 |
0.1460815607 |
- |
- |
PREDICTED: recQ-mediated genome instability protein 1 [Jatropha curcas] |
| 9 |
Hb_000260_770 |
0.1466759978 |
- |
- |
er lumen protein retaining receptor, putative [Ricinus communis] |
| 10 |
Hb_000134_250 |
0.1466910762 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_000679_110 |
0.1482540253 |
- |
- |
PREDICTED: lysM domain receptor-like kinase 3 [Jatropha curcas] |
| 12 |
Hb_000678_010 |
0.1492049417 |
- |
- |
PREDICTED: ketol-acid reductoisomerase, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_009659_030 |
0.1502880131 |
- |
- |
PREDICTED: beta-galactosidase 17 [Jatropha curcas] |
| 14 |
Hb_000229_050 |
0.1512147946 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 15 |
Hb_008053_060 |
0.1518142694 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |
| 16 |
Hb_001369_470 |
0.1530395508 |
- |
- |
PREDICTED: monocopper oxidase-like protein SKS1 [Jatropha curcas] |
| 17 |
Hb_007576_140 |
0.153570318 |
rubber biosynthesis |
Gene Name: 2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase |
2-C-methyl-D-erythritol 4-phosphate cytidylyltransferase [Hevea brasiliensis] |
| 18 |
Hb_005725_230 |
0.1555011332 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 19 |
Hb_000890_160 |
0.1558332863 |
- |
- |
PREDICTED: anaphase-promoting complex subunit 7 [Jatropha curcas] |
| 20 |
Hb_001195_220 |
0.1574265254 |
- |
- |
sugar transporter, putative [Ricinus communis] |