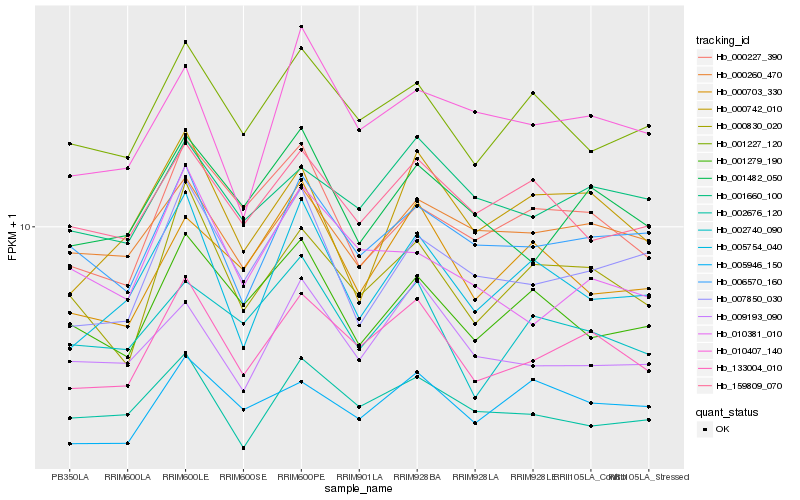
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_133004_010 |
0.0 |
- |
- |
PREDICTED: cyclin-D3-3 [Jatropha curcas] |
| 2 |
Hb_000830_020 |
0.0875988717 |
- |
- |
PREDICTED: 7-methylguanosine phosphate-specific 5'-nucleotidase A isoform X2 [Jatropha curcas] |
| 3 |
Hb_001482_050 |
0.0981506834 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 4 |
Hb_001660_100 |
0.1005862389 |
- |
- |
PREDICTED: glyoxylate reductase/hydroxypyruvate reductase [Jatropha curcas] |
| 5 |
Hb_006570_160 |
0.1023163661 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 6 |
Hb_001227_120 |
0.1053652082 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 7 |
Hb_005754_040 |
0.1093905219 |
- |
- |
PREDICTED: uncharacterized protein LOC105636678 isoform X2 [Jatropha curcas] |
| 8 |
Hb_009193_090 |
0.1119007234 |
- |
- |
PREDICTED: protein root UVB sensitive 3 isoform X1 [Jatropha curcas] |
| 9 |
Hb_000260_470 |
0.1132469561 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 10 |
Hb_000227_390 |
0.1138853327 |
- |
- |
PREDICTED: vacuole membrane protein KMS1 isoform X2 [Jatropha curcas] |
| 11 |
Hb_002676_120 |
0.1141541013 |
- |
- |
hypothetical protein POPTR_0002s23750g [Populus trichocarpa] |
| 12 |
Hb_005946_150 |
0.1146996895 |
- |
- |
PREDICTED: DNA mismatch repair protein MSH7 [Jatropha curcas] |
| 13 |
Hb_002740_090 |
0.1149245422 |
- |
- |
PREDICTED: putative membrane-bound O-acyltransferase C24H6.01c [Jatropha curcas] |
| 14 |
Hb_007850_030 |
0.1155737574 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 15 |
Hb_000742_010 |
0.1165565407 |
- |
- |
PREDICTED: ferredoxin--NADP reductase, root-type isozyme, chloroplastic isoform X2 [Jatropha curcas] |
| 16 |
Hb_159809_070 |
0.1178567118 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 17 |
Hb_000703_330 |
0.1187716665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 18 |
Hb_010407_140 |
0.1190802852 |
- |
- |
PREDICTED: malate dehydrogenase [Jatropha curcas] |
| 19 |
Hb_001279_190 |
0.1203287548 |
- |
- |
PREDICTED: uncharacterized membrane protein At3g27390 isoform X1 [Jatropha curcas] |
| 20 |
Hb_010381_010 |
0.1221450942 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |