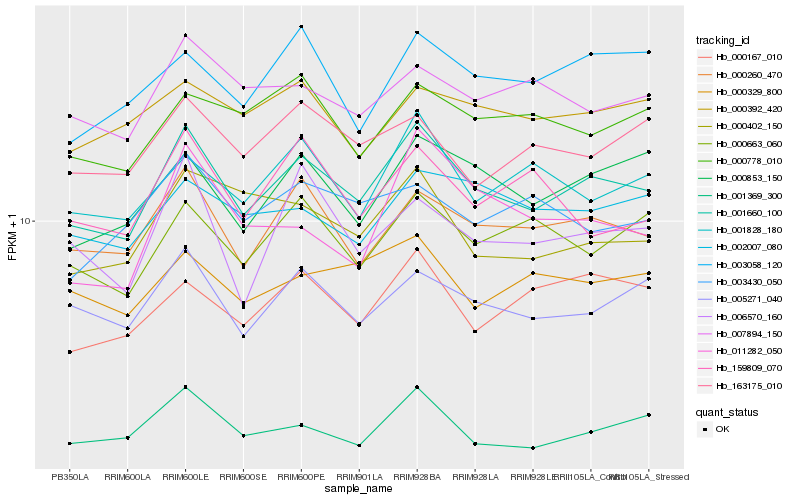
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001660_100 |
0.0 |
- |
- |
PREDICTED: glyoxylate reductase/hydroxypyruvate reductase [Jatropha curcas] |
| 2 |
Hb_000260_470 |
0.0655617343 |
- |
- |
PREDICTED: uncharacterized protein LOC105649044 [Jatropha curcas] |
| 3 |
Hb_005271_040 |
0.0750028226 |
- |
- |
PREDICTED: serine racemase [Jatropha curcas] |
| 4 |
Hb_000853_150 |
0.0779172394 |
- |
- |
Fumarase 1 isoform 2 [Theobroma cacao] |
| 5 |
Hb_001369_300 |
0.0782072934 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 2-1-like [Jatropha curcas] |
| 6 |
Hb_000663_060 |
0.083261176 |
- |
- |
hypothetical protein JCGZ_16277 [Jatropha curcas] |
| 7 |
Hb_001828_180 |
0.084479656 |
- |
- |
26S proteasome regulatory subunit S3, putative [Ricinus communis] |
| 8 |
Hb_000167_010 |
0.0862652714 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000402_150 |
0.0868811546 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 10 |
Hb_007894_150 |
0.087613818 |
- |
- |
hypothetical protein EUGRSUZ_F03462 [Eucalyptus grandis] |
| 11 |
Hb_002007_080 |
0.0882765404 |
- |
- |
PREDICTED: acyl-CoA-binding domain-containing protein 4-like [Jatropha curcas] |
| 12 |
Hb_000329_800 |
0.0885277407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_000778_010 |
0.0889986974 |
- |
- |
hypothetical protein [Bacillus subtilis] |
| 14 |
Hb_003058_120 |
0.0891869609 |
- |
- |
PREDICTED: 26S proteasome non-ATPase regulatory subunit 4 homolog [Jatropha curcas] |
| 15 |
Hb_163175_010 |
0.090009391 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 16 |
Hb_006570_160 |
0.0926179225 |
- |
- |
PREDICTED: putative ER lumen protein-retaining receptor C28H8.4 [Jatropha curcas] |
| 17 |
Hb_000392_420 |
0.0928324644 |
- |
- |
PREDICTED: probable aldehyde dehydrogenase isoform X1 [Jatropha curcas] |
| 18 |
Hb_159809_070 |
0.0939174456 |
- |
- |
phospholipase A-2-activating protein, putative [Ricinus communis] |
| 19 |
Hb_003430_050 |
0.0947080694 |
- |
- |
PREDICTED: serine/arginine-rich splicing factor RSZ21A isoform X1 [Phoenix dactylifera] |
| 20 |
Hb_011282_050 |
0.0956412033 |
- |
- |
PREDICTED: diaminopimelate decarboxylase 2, chloroplastic-like [Jatropha curcas] |