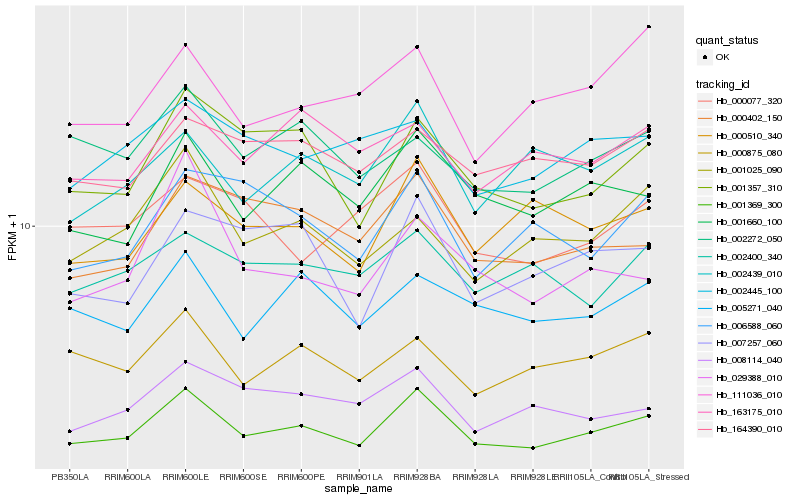
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_300 |
0.0 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 2-1-like [Jatropha curcas] |
| 2 |
Hb_001660_100 |
0.0782072934 |
- |
- |
PREDICTED: glyoxylate reductase/hydroxypyruvate reductase [Jatropha curcas] |
| 3 |
Hb_001357_310 |
0.08282966 |
- |
- |
PREDICTED: dihydrolipoyllysine-residue succinyltransferase component of 2-oxoglutarate dehydrogenase complex 2, mitochondrial-like [Jatropha curcas] |
| 4 |
Hb_000510_340 |
0.0903803288 |
transcription factor |
TF Family: bZIP |
PREDICTED: light-inducible protein CPRF2 [Jatropha curcas] |
| 5 |
Hb_000402_150 |
0.0908082196 |
- |
- |
PREDICTED: uncharacterized protein LOC105644650 isoform X2 [Jatropha curcas] |
| 6 |
Hb_005271_040 |
0.0910865054 |
- |
- |
PREDICTED: serine racemase [Jatropha curcas] |
| 7 |
Hb_000875_080 |
0.09139835 |
- |
- |
PREDICTED: (+)-delta-cadinene synthase isozyme XC14-like isoform X2 [Gossypium raimondii] |
| 8 |
Hb_002439_010 |
0.0934705431 |
- |
- |
PREDICTED: DNA damage-inducible protein 1 [Jatropha curcas] |
| 9 |
Hb_007257_060 |
0.0943475552 |
- |
- |
PREDICTED: omega-amidase,chloroplastic [Jatropha curcas] |
| 10 |
Hb_008114_040 |
0.0947436607 |
- |
- |
Fanconi anemia group D2 [Gossypium arboreum] |
| 11 |
Hb_001025_090 |
0.0969771392 |
- |
- |
PREDICTED: FKBP12-interacting protein of 37 kDa [Jatropha curcas] |
| 12 |
Hb_002400_340 |
0.0999421123 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 13 |
Hb_006588_060 |
0.100494211 |
- |
- |
PREDICTED: DDB1- and CUL4-associated factor 13 [Jatropha curcas] |
| 14 |
Hb_002272_050 |
0.1008775203 |
- |
- |
microsomal signal peptidase 23 kD subunit, putative [Ricinus communis] |
| 15 |
Hb_164390_010 |
0.1012878896 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor HBP-1a [Jatropha curcas] |
| 16 |
Hb_000077_320 |
0.1015201021 |
- |
- |
PREDICTED: lanC-like protein GCL2 [Jatropha curcas] |
| 17 |
Hb_002445_100 |
0.1017542155 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 5 isoform X2 [Jatropha curcas] |
| 18 |
Hb_111036_010 |
0.1018520882 |
- |
- |
Eukaryotic translation initiation factor 6-2 [Glycine soja] |
| 19 |
Hb_163175_010 |
0.1028814613 |
- |
- |
hypothetical protein CISIN_1g036035mg, partial [Citrus sinensis] |
| 20 |
Hb_029388_010 |
0.1034483134 |
transcription factor |
TF Family: C2C2-CO-like |
PREDICTED: zinc finger protein CONSTANS-LIKE 15-like [Jatropha curcas] |