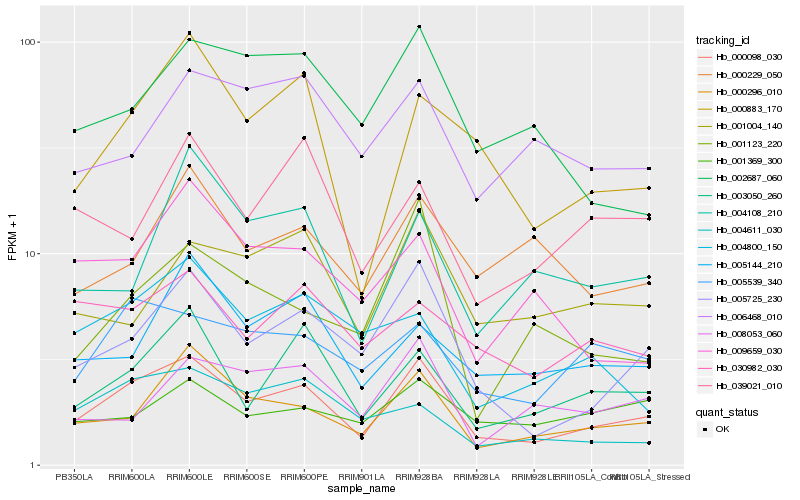
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000098_030 |
0.0 |
- |
- |
MLO-like protein 4 [Gossypium arboreum] |
| 2 |
Hb_005725_230 |
0.1234705739 |
transcription factor |
TF Family: AP2 |
PREDICTED: AP2-like ethylene-responsive transcription factor ANT [Jatropha curcas] |
| 3 |
Hb_000296_010 |
0.1291968898 |
- |
- |
PREDICTED: TSL-kinase interacting protein 1 [Jatropha curcas] |
| 4 |
Hb_003050_260 |
0.1474952311 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 5 |
Hb_000883_170 |
0.1489594557 |
- |
- |
senescence-associated family protein [Populus trichocarpa] |
| 6 |
Hb_005539_340 |
0.1574316354 |
- |
- |
PREDICTED: CDT1-like protein a, chloroplastic [Jatropha curcas] |
| 7 |
Hb_005144_210 |
0.1688628 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 8 |
Hb_001004_140 |
0.1704489129 |
- |
- |
hypothetical protein JCGZ_26893 [Jatropha curcas] |
| 9 |
Hb_030982_030 |
0.1715940775 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000229_050 |
0.1742853282 |
- |
- |
PREDICTED: protein STRUBBELIG-RECEPTOR FAMILY 3-like [Jatropha curcas] |
| 11 |
Hb_004611_030 |
0.176492475 |
- |
- |
structural maintenance of chromosomes 5 smc5, putative [Ricinus communis] |
| 12 |
Hb_039021_010 |
0.1767148948 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 13 |
Hb_004800_150 |
0.1767254925 |
transcription factor |
TF Family: SNF2 |
PREDICTED: ATP-dependent DNA helicase DDM1 [Jatropha curcas] |
| 14 |
Hb_004108_210 |
0.1768481745 |
- |
- |
phenazine biosynthesis protein, putative [Ricinus communis] |
| 15 |
Hb_002687_060 |
0.1781834574 |
- |
- |
PREDICTED: pyruvate kinase, cytosolic isozyme [Jatropha curcas] |
| 16 |
Hb_006468_010 |
0.1791026187 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_001369_300 |
0.179519598 |
- |
- |
PREDICTED: structural maintenance of chromosomes protein 2-1-like [Jatropha curcas] |
| 18 |
Hb_001123_220 |
0.1798585359 |
- |
- |
- |
| 19 |
Hb_008053_060 |
0.1800500302 |
- |
- |
PREDICTED: protein PAIR1 isoform X2 [Jatropha curcas] |
| 20 |
Hb_009659_030 |
0.1807590433 |
- |
- |
PREDICTED: beta-galactosidase 17 [Jatropha curcas] |