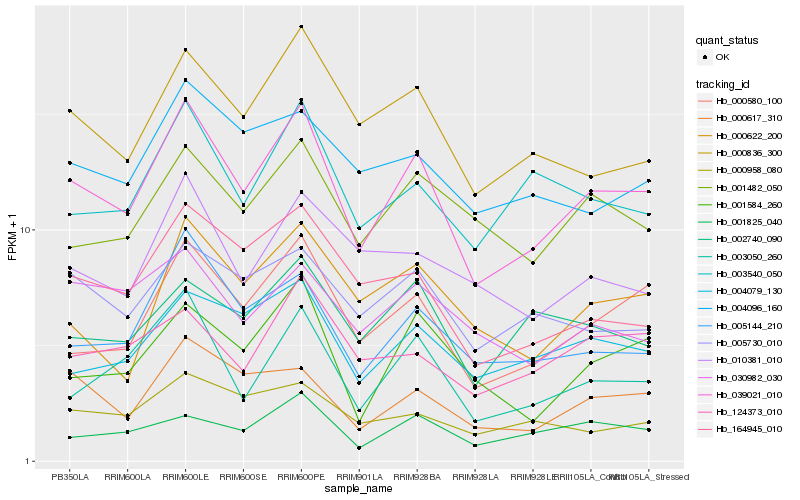
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_039021_010 |
0.0 |
transcription factor |
TF Family: B3 |
hypothetical protein RCOM_0465210 [Ricinus communis] |
| 2 |
Hb_000580_100 |
0.0972571065 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 3 |
Hb_005144_210 |
0.1095580519 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_000836_300 |
0.1122630237 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 5 |
Hb_004079_130 |
0.1159352068 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 6 |
Hb_003050_260 |
0.115983008 |
- |
- |
DNA replication licensing factor MCM3, putative [Ricinus communis] |
| 7 |
Hb_000617_310 |
0.1173227185 |
- |
- |
PREDICTED: cell cycle checkpoint control protein RAD9A [Jatropha curcas] |
| 8 |
Hb_164945_010 |
0.1197581843 |
- |
- |
PREDICTED: DNA primase small subunit [Jatropha curcas] |
| 9 |
Hb_003540_050 |
0.120307563 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 10 |
Hb_001825_040 |
0.1232969478 |
- |
- |
myosin XI, putative [Ricinus communis] |
| 11 |
Hb_005730_010 |
0.1238411312 |
- |
- |
PREDICTED: ubiquitin carboxyl-terminal hydrolase 7 isoform X1 [Populus euphratica] |
| 12 |
Hb_001482_050 |
0.1244491523 |
- |
- |
COR413-PM2, putative [Ricinus communis] |
| 13 |
Hb_001584_260 |
0.1255299441 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 14 |
Hb_000622_200 |
0.1286332378 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 15 |
Hb_124373_010 |
0.1291087158 |
- |
- |
PREDICTED: BRCA1-A complex subunit BRE isoform X1 [Jatropha curcas] |
| 16 |
Hb_000958_080 |
0.1296606399 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 17 |
Hb_002740_090 |
0.129742686 |
- |
- |
PREDICTED: putative membrane-bound O-acyltransferase C24H6.01c [Jatropha curcas] |
| 18 |
Hb_004096_160 |
0.1304571726 |
- |
- |
PREDICTED: T-complex protein 1 subunit zeta 1 [Jatropha curcas] |
| 19 |
Hb_010381_010 |
0.1310772447 |
- |
- |
WD-repeat protein, putative [Ricinus communis] |
| 20 |
Hb_030982_030 |
0.1322992088 |
- |
- |
PREDICTED: uncharacterized protein LOC105636001 isoform X1 [Jatropha curcas] |