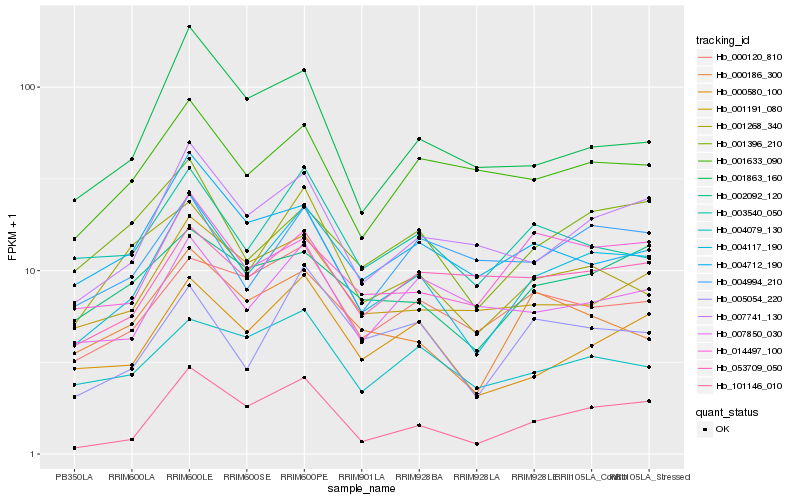
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004117_190 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105649109 [Jatropha curcas] |
| 2 |
Hb_000580_100 |
0.1101791875 |
- |
- |
PREDICTED: uncharacterized protein LOC105643133 isoform X1 [Jatropha curcas] |
| 3 |
Hb_007741_130 |
0.1108019892 |
- |
- |
hypothetical protein B456_007G089000 [Gossypium raimondii] |
| 4 |
Hb_005054_220 |
0.118244106 |
- |
- |
PREDICTED: peroxisomal nicotinamide adenine dinucleotide carrier isoform X1 [Jatropha curcas] |
| 5 |
Hb_101146_010 |
0.1191553455 |
- |
- |
PREDICTED: peregrin-like [Jatropha curcas] |
| 6 |
Hb_003540_050 |
0.1223512428 |
- |
- |
PREDICTED: plant intracellular Ras-group-related LRR protein 6 isoform X4 [Jatropha curcas] |
| 7 |
Hb_001268_340 |
0.1247383813 |
- |
- |
Actin, putative [Ricinus communis] |
| 8 |
Hb_001396_210 |
0.1272530366 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_000120_810 |
0.1277376904 |
- |
- |
serine/threonine-protein kinase cx32, putative [Ricinus communis] |
| 10 |
Hb_001191_080 |
0.1286912435 |
- |
- |
protein with unknown function [Ricinus communis] |
| 11 |
Hb_000186_300 |
0.1305262985 |
- |
- |
PREDICTED: malonyl-CoA decarboxylase, mitochondrial [Jatropha curcas] |
| 12 |
Hb_007850_030 |
0.1306698744 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |
| 13 |
Hb_004079_130 |
0.1335658505 |
- |
- |
PREDICTED: uncharacterized protein LOC105637708 [Jatropha curcas] |
| 14 |
Hb_002092_120 |
0.1342999919 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase, chloroplastic [Jatropha curcas] |
| 15 |
Hb_014497_100 |
0.1351645073 |
- |
- |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 16 |
Hb_004712_190 |
0.1371825174 |
- |
- |
PREDICTED: uncharacterized protein LOC105633105 [Jatropha curcas] |
| 17 |
Hb_001633_090 |
0.1376610017 |
- |
- |
PREDICTED: mitochondrial phosphate carrier protein 3, mitochondrial [Jatropha curcas] |
| 18 |
Hb_004994_210 |
0.1397255492 |
- |
- |
phosphomannomutase family protein [Populus trichocarpa] |
| 19 |
Hb_001863_160 |
0.1402920516 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 20 |
Hb_053709_050 |
0.1416427163 |
- |
- |
PREDICTED: nudix hydrolase 23, chloroplastic isoform X2 [Jatropha curcas] |