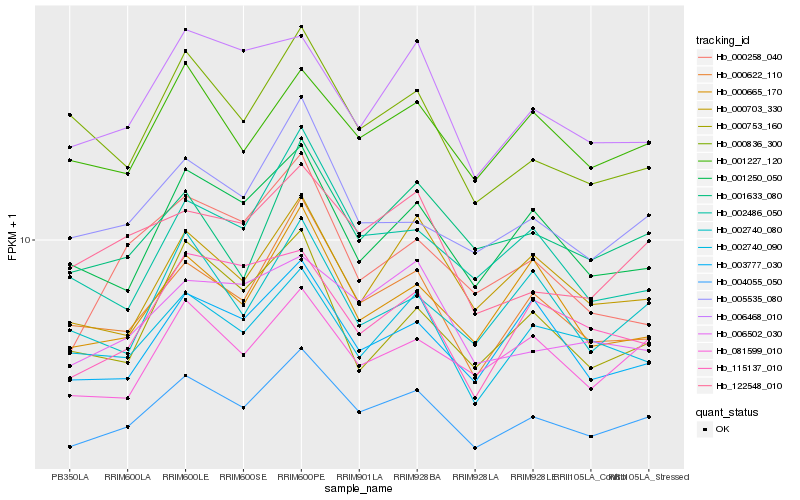
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004055_050 |
0.0 |
- |
- |
PREDICTED: coatomer subunit zeta-1-like [Jatropha curcas] |
| 2 |
Hb_000622_110 |
0.0797409822 |
- |
- |
cmp-sialic acid transporter, putative [Ricinus communis] |
| 3 |
Hb_001250_050 |
0.0959819371 |
- |
- |
PREDICTED: uncharacterized protein LOC105644623 isoform X1 [Jatropha curcas] |
| 4 |
Hb_005535_080 |
0.0999414813 |
- |
- |
hypothetical protein CICLE_v10012766mg [Citrus clementina] |
| 5 |
Hb_081599_010 |
0.1004015848 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_003777_030 |
0.1005833114 |
- |
- |
Exocyst complex component, putative [Ricinus communis] |
| 7 |
Hb_000665_170 |
0.1042067746 |
- |
- |
PREDICTED: probable beta-1,3-galactosyltransferase 10 [Jatropha curcas] |
| 8 |
Hb_002740_090 |
0.1044284218 |
- |
- |
PREDICTED: putative membrane-bound O-acyltransferase C24H6.01c [Jatropha curcas] |
| 9 |
Hb_000703_330 |
0.1049716372 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_122548_010 |
0.1059675304 |
- |
- |
acyl-CoA oxidase, putative [Ricinus communis] |
| 11 |
Hb_002486_050 |
0.1084936074 |
- |
- |
Multiple inositol polyphosphate phosphatase 1 precursor, putative [Ricinus communis] |
| 12 |
Hb_000836_300 |
0.1091938878 |
- |
- |
caax prenyl protease ste24, putative [Ricinus communis] |
| 13 |
Hb_006502_030 |
0.109433232 |
- |
- |
PREDICTED: probable kinetochore protein ndc80 [Jatropha curcas] |
| 14 |
Hb_001227_120 |
0.1095798712 |
transcription factor |
TF Family: C2H2 |
Histone deacetylase 2a, putative [Ricinus communis] |
| 15 |
Hb_001633_080 |
0.1099485324 |
- |
- |
PREDICTED: V-type proton ATPase subunit C [Gossypium raimondii] |
| 16 |
Hb_006468_010 |
0.1105870224 |
- |
- |
PREDICTED: eukaryotic translation initiation factor isoform 4G-1 isoform X1 [Jatropha curcas] |
| 17 |
Hb_115137_010 |
0.1107722854 |
- |
- |
PREDICTED: probable protein S-acyltransferase 23 isoform X2 [Jatropha curcas] |
| 18 |
Hb_002740_080 |
0.1112238724 |
- |
- |
flap endonuclease-1, putative [Ricinus communis] |
| 19 |
Hb_000258_040 |
0.112522262 |
- |
- |
PREDICTED: probable receptor-like protein kinase At3g55450 [Jatropha curcas] |
| 20 |
Hb_000753_160 |
0.1137485553 |
- |
- |
PREDICTED: uncharacterized protein LOC105640669 [Jatropha curcas] |