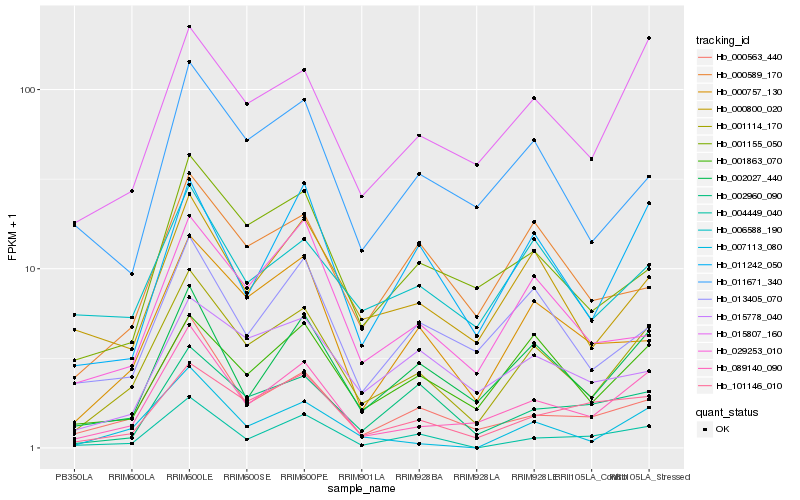
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001114_170 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_089140_090 |
0.1198407624 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X3 [Beta vulgaris subsp. vulgaris] |
| 3 |
Hb_000757_130 |
0.1398092952 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 4 |
Hb_015807_160 |
0.1398987523 |
- |
- |
PREDICTED: probable histone H2A.1 [Jatropha curcas] |
| 5 |
Hb_002027_440 |
0.1426411521 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_001863_070 |
0.1435354528 |
- |
- |
Queuine tRNA-ribosyltransferase [Theobroma cacao] |
| 7 |
Hb_101146_010 |
0.1466455332 |
- |
- |
PREDICTED: peregrin-like [Jatropha curcas] |
| 8 |
Hb_000800_020 |
0.148574693 |
- |
- |
PREDICTED: 3-phosphoinositide-dependent protein kinase 2 [Jatropha curcas] |
| 9 |
Hb_001155_050 |
0.1506147124 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 10 |
Hb_000563_440 |
0.1513238545 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 11 |
Hb_011242_050 |
0.1571442036 |
- |
- |
PREDICTED: ubiquitin-conjugating enzyme E2-23 kDa [Jatropha curcas] |
| 12 |
Hb_006588_190 |
0.1620206758 |
- |
- |
PREDICTED: ataxin-3 homolog [Jatropha curcas] |
| 13 |
Hb_007113_080 |
0.1622320138 |
- |
- |
PREDICTED: uncharacterized protein LOC105639553 isoform X1 [Jatropha curcas] |
| 14 |
Hb_013405_070 |
0.1629692981 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 15 |
Hb_011671_340 |
0.1630073777 |
- |
- |
small GTPase [Hevea brasiliensis] |
| 16 |
Hb_029253_010 |
0.165747855 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 17 |
Hb_002960_090 |
0.1671912774 |
- |
- |
protein with unknown function [Ricinus communis] |
| 18 |
Hb_000589_170 |
0.1689379903 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 19 |
Hb_015778_040 |
0.1703390406 |
- |
- |
PREDICTED: uncharacterized protein LOC105650695 [Jatropha curcas] |
| 20 |
Hb_004449_040 |
0.1706983711 |
- |
- |
PREDICTED: uncharacterized protein C630.12 [Jatropha curcas] |