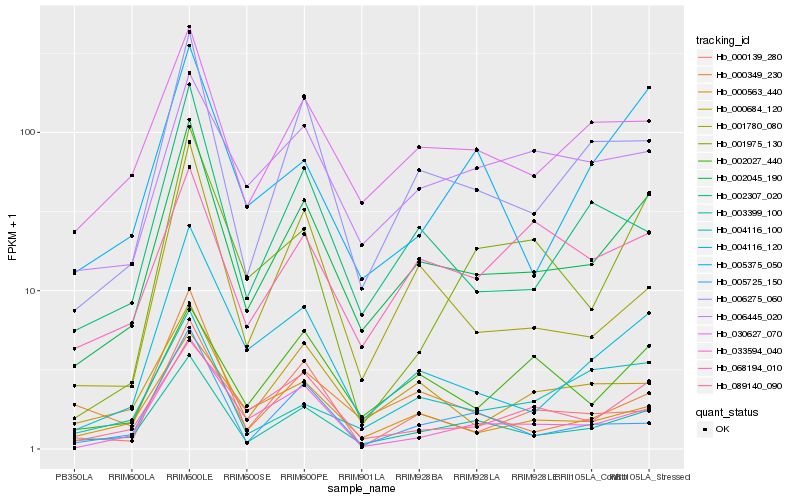
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002045_190 |
0.0 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 2 |
Hb_003399_100 |
0.0997896767 |
- |
- |
PREDICTED: putative cyclin-A3-1 [Jatropha curcas] |
| 3 |
Hb_006275_060 |
0.1209985517 |
- |
- |
hypothetical protein CARUB_v10021660mg, partial [Capsella rubella] |
| 4 |
Hb_004116_120 |
0.1338200411 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 5 |
Hb_030627_070 |
0.1354872877 |
- |
- |
histone H4 [Zea mays] |
| 6 |
Hb_089140_090 |
0.1479032694 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X3 [Beta vulgaris subsp. vulgaris] |
| 7 |
Hb_033594_040 |
0.1544466337 |
- |
- |
PREDICTED: heptahelical transmembrane protein 4-like isoform X2 [Jatropha curcas] |
| 8 |
Hb_000563_440 |
0.1550075569 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 9 |
Hb_002307_020 |
0.1629970217 |
- |
- |
PREDICTED: histone H1-like [Jatropha curcas] |
| 10 |
Hb_000684_120 |
0.1658680289 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 11 |
Hb_000139_280 |
0.1725944393 |
- |
- |
PREDICTED: mitogen-activated protein kinase kinase kinase NPK1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_001780_080 |
0.1757970192 |
- |
- |
PREDICTED: tubulin beta-2 chain [Jatropha curcas] |
| 13 |
Hb_004116_100 |
0.1763579616 |
- |
- |
Isoamyl acetate-hydrolyzing esterase, putative [Ricinus communis] |
| 14 |
Hb_005375_050 |
0.1791834765 |
- |
- |
inorganic pyrophosphatase, putative [Ricinus communis] |
| 15 |
Hb_002027_440 |
0.1802133876 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 16 |
Hb_006445_020 |
0.1823473157 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |
| 17 |
Hb_001975_130 |
0.1828376577 |
- |
- |
PREDICTED: probable galactinol--sucrose galactosyltransferase 6 isoform X1 [Populus euphratica] |
| 18 |
Hb_005725_150 |
0.1847837777 |
- |
- |
PREDICTED: kinesin-related protein 11 isoform X1 [Jatropha curcas] |
| 19 |
Hb_068194_010 |
0.1870370552 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 20 |
Hb_000349_230 |
0.1873318564 |
- |
- |
PREDICTED: protein TPX2 [Jatropha curcas] |