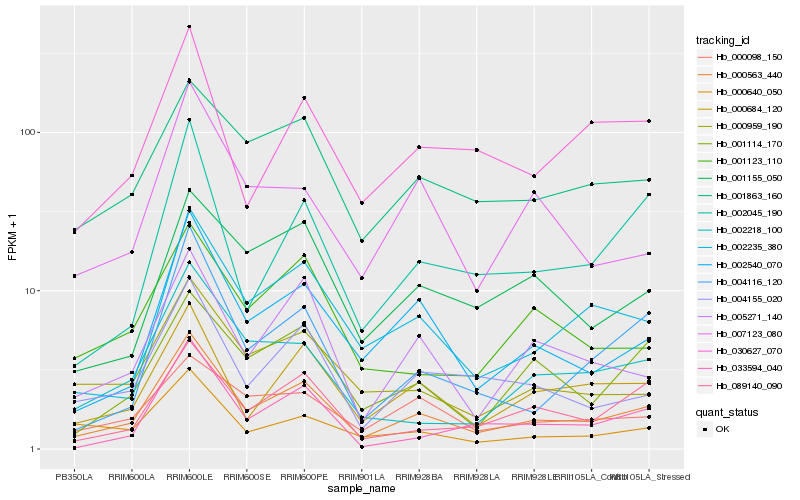
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000563_440 |
0.0 |
- |
- |
PREDICTED: condensin-2 complex subunit H2 isoform X2 [Jatropha curcas] |
| 2 |
Hb_002540_070 |
0.1233955123 |
- |
- |
PREDICTED: protein cornichon homolog 1 [Jatropha curcas] |
| 3 |
Hb_000684_120 |
0.124074519 |
- |
- |
PREDICTED: uncharacterized protein LOC105642190 isoform X2 [Jatropha curcas] |
| 4 |
Hb_002235_380 |
0.1321746433 |
- |
- |
hypothetical protein JCGZ_19860 [Jatropha curcas] |
| 5 |
Hb_004155_020 |
0.1335246435 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 6 |
Hb_005271_140 |
0.1408478404 |
transcription factor |
TF Family: LIM |
Pollen-specific protein SF3 [Morus notabilis] |
| 7 |
Hb_001863_160 |
0.1417060778 |
- |
- |
PREDICTED: 14-3-3-like protein [Jatropha curcas] |
| 8 |
Hb_000959_190 |
0.1423865802 |
- |
- |
PREDICTED: crossover junction endonuclease EME1B-like [Jatropha curcas] |
| 9 |
Hb_089140_090 |
0.143878288 |
- |
- |
PREDICTED: ankyrin repeat-containing protein At3g12360-like isoform X3 [Beta vulgaris subsp. vulgaris] |
| 10 |
Hb_004116_120 |
0.1505268436 |
- |
- |
PREDICTED: protein EARLY FLOWERING 4 [Jatropha curcas] |
| 11 |
Hb_030627_070 |
0.1511567922 |
- |
- |
histone H4 [Zea mays] |
| 12 |
Hb_001114_170 |
0.1513238545 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 13 |
Hb_002218_100 |
0.1517799765 |
- |
- |
PREDICTED: uncharacterized protein At2g34160 [Jatropha curcas] |
| 14 |
Hb_000098_150 |
0.1526901446 |
- |
- |
DNA helicase hus2, putative [Ricinus communis] |
| 15 |
Hb_002045_190 |
0.1550075569 |
- |
- |
PREDICTED: histone H2A variant 1 [Jatropha curcas] |
| 16 |
Hb_001155_050 |
0.1552224675 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 17 |
Hb_033594_040 |
0.1554749702 |
- |
- |
PREDICTED: heptahelical transmembrane protein 4-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000640_050 |
0.1573599814 |
- |
- |
DNA replication licensing factor MCM7, putative [Ricinus communis] |
| 19 |
Hb_001123_110 |
0.1581561877 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007123_080 |
0.1604752493 |
- |
- |
big map kinase/bmk, putative [Ricinus communis] |