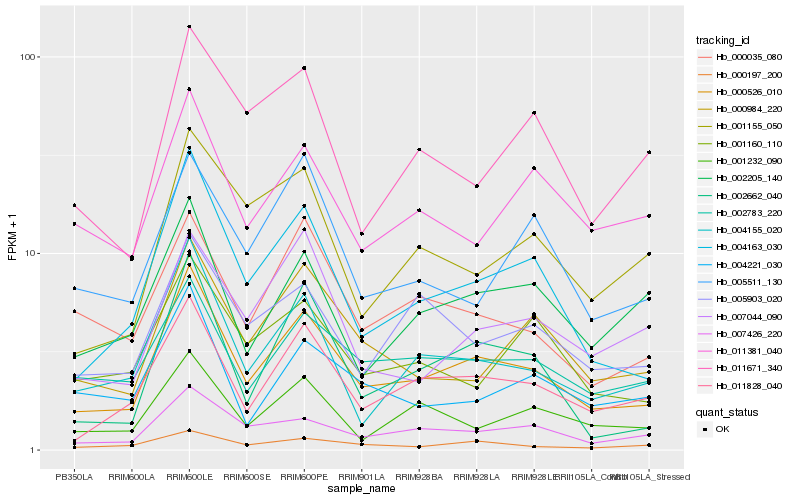
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000526_010 |
0.0 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 2 |
Hb_004163_030 |
0.0974294778 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 3 |
Hb_011828_040 |
0.1137354572 |
- |
- |
PREDICTED: uncharacterized protein LOC105642173 isoform X2 [Jatropha curcas] |
| 4 |
Hb_007426_220 |
0.1243885635 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 5 |
Hb_004155_020 |
0.1251365983 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 6 |
Hb_002783_220 |
0.1319838236 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 7 |
Hb_002662_040 |
0.1377324521 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 8 |
Hb_000984_220 |
0.1390778278 |
- |
- |
PREDICTED: inositol-tetrakisphosphate 1-kinase 2-like [Populus euphratica] |
| 9 |
Hb_000197_200 |
0.1413927816 |
- |
- |
Sec14 cytosolic factor, putative [Ricinus communis] |
| 10 |
Hb_000035_080 |
0.1492660974 |
- |
- |
PREDICTED: U-box domain-containing protein 12-like [Jatropha curcas] |
| 11 |
Hb_005903_020 |
0.1496071252 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 12 |
Hb_002205_140 |
0.1530921976 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 13 |
Hb_001160_110 |
0.1554391447 |
- |
- |
PREDICTED: kinesin-related protein 13 [Jatropha curcas] |
| 14 |
Hb_001155_050 |
0.1566770335 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 15 |
Hb_004221_030 |
0.1569373251 |
transcription factor |
TF Family: bHLH |
PREDICTED: transcription factor SPATULA-like isoform X1 [Jatropha curcas] |
| 16 |
Hb_001232_090 |
0.1569899058 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 17 |
Hb_007044_090 |
0.1596038303 |
- |
- |
PREDICTED: isoprenylcysteine alpha-carbonyl methylesterase ICME isoform X2 [Jatropha curcas] |
| 18 |
Hb_011381_040 |
0.1609675406 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 19 |
Hb_011671_340 |
0.1639463245 |
- |
- |
small GTPase [Hevea brasiliensis] |
| 20 |
Hb_005511_130 |
0.1649876444 |
- |
- |
PREDICTED: transmembrane protein 115-like [Populus euphratica] |