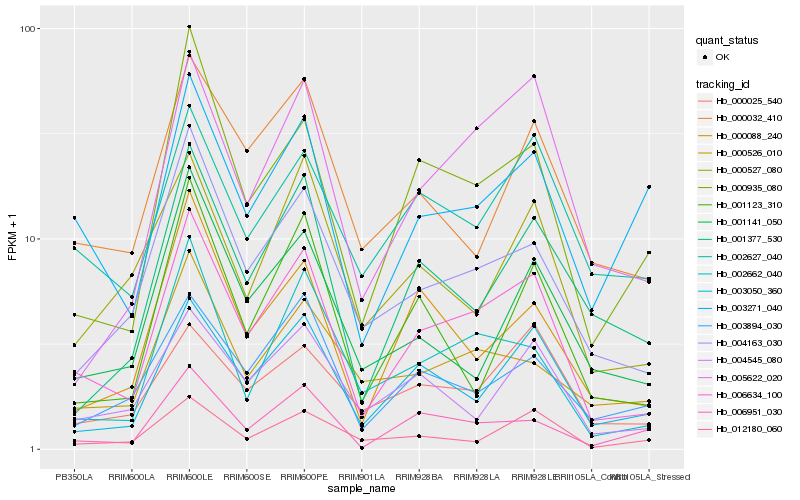
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_006634_100 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000935_080 |
0.1377469098 |
- |
- |
PREDICTED: branched-chain-amino-acid aminotransferase 3, chloroplastic isoform X1 [Jatropha curcas] |
| 3 |
Hb_004163_030 |
0.13779453 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 4 |
Hb_003050_360 |
0.1409488746 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 5 |
Hb_005622_020 |
0.1448015453 |
- |
- |
PREDICTED: DCC family protein At1g52590, chloroplastic [Jatropha curcas] |
| 6 |
Hb_002662_040 |
0.1493556946 |
- |
- |
PREDICTED: uncharacterized protein LOC105630915 [Jatropha curcas] |
| 7 |
Hb_006951_030 |
0.1577818835 |
- |
- |
PREDICTED: protein MCM10 homolog [Jatropha curcas] |
| 8 |
Hb_000088_240 |
0.1592149585 |
transcription factor |
TF Family: MYB |
r2r3-myb transcription factor, putative [Ricinus communis] |
| 9 |
Hb_001377_530 |
0.1611353995 |
- |
- |
NC domain-containing family protein [Populus trichocarpa] |
| 10 |
Hb_000032_410 |
0.1617772082 |
- |
- |
PREDICTED: dynamin-related protein 5A [Jatropha curcas] |
| 11 |
Hb_003894_030 |
0.1641720493 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 12 |
Hb_002627_040 |
0.1644283274 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 13 |
Hb_004545_080 |
0.1644433259 |
- |
- |
protein with unknown function [Ricinus communis] |
| 14 |
Hb_003271_040 |
0.1691111323 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |
| 15 |
Hb_000527_080 |
0.1692318849 |
- |
- |
PREDICTED: caffeoylshikimate esterase [Jatropha curcas] |
| 16 |
Hb_000526_010 |
0.1717735412 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 17 |
Hb_001123_310 |
0.1723835519 |
- |
- |
PREDICTED: coiled-coil domain-containing protein SCD2 [Jatropha curcas] |
| 18 |
Hb_001141_050 |
0.1724838126 |
transcription factor |
TF Family: CPP |
PREDICTED: protein tesmin/TSO1-like CXC 2 isoform X2 [Jatropha curcas] |
| 19 |
Hb_000025_540 |
0.1730230396 |
- |
- |
PREDICTED: uncharacterized protein LOC104879644 [Vitis vinifera] |
| 20 |
Hb_012180_060 |
0.1741653269 |
- |
- |
peptidase, putative [Ricinus communis] |