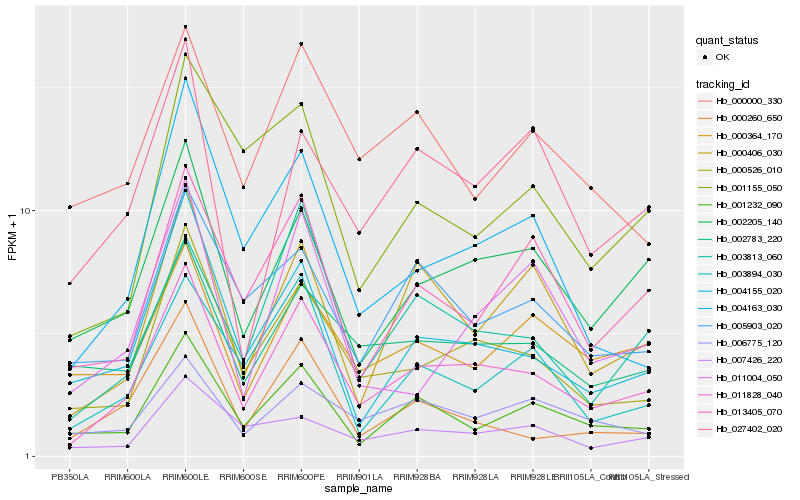
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_011828_040 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105642173 isoform X2 [Jatropha curcas] |
| 2 |
Hb_000526_010 |
0.1137354572 |
- |
- |
PREDICTED: uroporphyrinogen-III C-methyltransferase [Jatropha curcas] |
| 3 |
Hb_002205_140 |
0.1311995176 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 4 |
Hb_004155_020 |
0.1413293811 |
transcription factor |
TF Family: MYB |
myb, putative [Ricinus communis] |
| 5 |
Hb_001232_090 |
0.144327977 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 6 |
Hb_003813_060 |
0.1444925361 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_000406_030 |
0.1457159854 |
- |
- |
Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase [Morus notabilis] |
| 8 |
Hb_004163_030 |
0.1476218368 |
- |
- |
PREDICTED: protein PHLOEM PROTEIN 2-LIKE A10 [Jatropha curcas] |
| 9 |
Hb_013405_070 |
0.1517139998 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 10 |
Hb_003894_030 |
0.1527940779 |
- |
- |
plant sec1, putative [Ricinus communis] |
| 11 |
Hb_002783_220 |
0.1531660463 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 12 |
Hb_005903_020 |
0.1598497496 |
- |
- |
12-oxophytodienoate reductase 2 [Theobroma cacao] |
| 13 |
Hb_007426_220 |
0.1606033666 |
- |
- |
Endoplasmic oxidoreductin-1 precursor, putative [Ricinus communis] |
| 14 |
Hb_000364_170 |
0.1621562243 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 15 |
Hb_027402_020 |
0.1629110705 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 16 |
Hb_006775_120 |
0.1636005349 |
- |
- |
exonuclease-like protein [Oryza sativa Japonica Group] |
| 17 |
Hb_000000_330 |
0.1662305349 |
- |
- |
PREDICTED: uncharacterized protein LOC105633747 [Jatropha curcas] |
| 18 |
Hb_000260_650 |
0.1677313928 |
- |
- |
PREDICTED: protein PFC0760c [Jatropha curcas] |
| 19 |
Hb_001155_050 |
0.1681497664 |
- |
- |
PREDICTED: KH domain-containing protein SPIN1-like [Jatropha curcas] |
| 20 |
Hb_011004_050 |
0.1684125605 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |