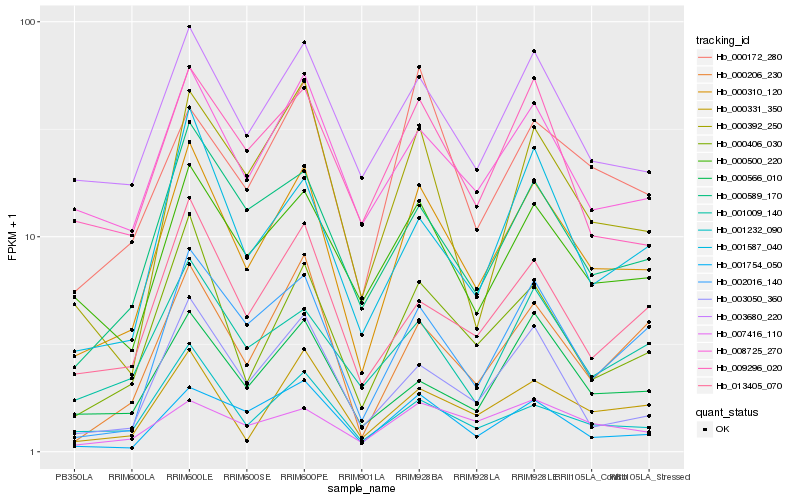
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000310_120 |
0.0 |
- |
- |
PREDICTED: probable transaldolase [Jatropha curcas] |
| 2 |
Hb_000406_030 |
0.104632375 |
- |
- |
Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase [Morus notabilis] |
| 3 |
Hb_003680_220 |
0.1141260763 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 4 |
Hb_000589_170 |
0.1141868111 |
- |
- |
PREDICTED: telomere repeat-binding protein 4 isoform X3 [Jatropha curcas] |
| 5 |
Hb_000500_220 |
0.1188521334 |
- |
- |
PREDICTED: intersectin-1 [Jatropha curcas] |
| 6 |
Hb_008725_270 |
0.1204817758 |
- |
- |
PREDICTED: probable protein disulfide-isomerase A6 [Jatropha curcas] |
| 7 |
Hb_013405_070 |
0.1218436411 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 8 |
Hb_002016_140 |
0.1232392633 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase BAH1-like 1 [Jatropha curcas] |
| 9 |
Hb_001587_040 |
0.1242057271 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 10 |
Hb_001232_090 |
0.1270856826 |
- |
- |
PREDICTED: regulator of telomere elongation helicase 1 homolog [Jatropha curcas] |
| 11 |
Hb_000331_350 |
0.1275823609 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 12 |
Hb_003050_360 |
0.1299002009 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1 isoform X2 [Jatropha curcas] |
| 13 |
Hb_001754_050 |
0.1304546309 |
- |
- |
protein arginine n-methyltransferase, putative [Ricinus communis] |
| 14 |
Hb_000566_010 |
0.1324429201 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 15 |
Hb_000392_250 |
0.1336816563 |
- |
- |
PREDICTED: HVA22-like protein a [Jatropha curcas] |
| 16 |
Hb_000172_280 |
0.133707405 |
- |
- |
aspartate aminotransferase, putative [Ricinus communis] |
| 17 |
Hb_009296_020 |
0.1339828809 |
desease resistance |
Gene Name: ATP-synt_ab_N |
PREDICTED: ATP synthase subunit beta, mitochondrial-like [Jatropha curcas] |
| 18 |
Hb_001009_140 |
0.1340380973 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 19 |
Hb_000206_230 |
0.1340999783 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_007416_110 |
0.1350339668 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 14 isoform X1 [Jatropha curcas] |