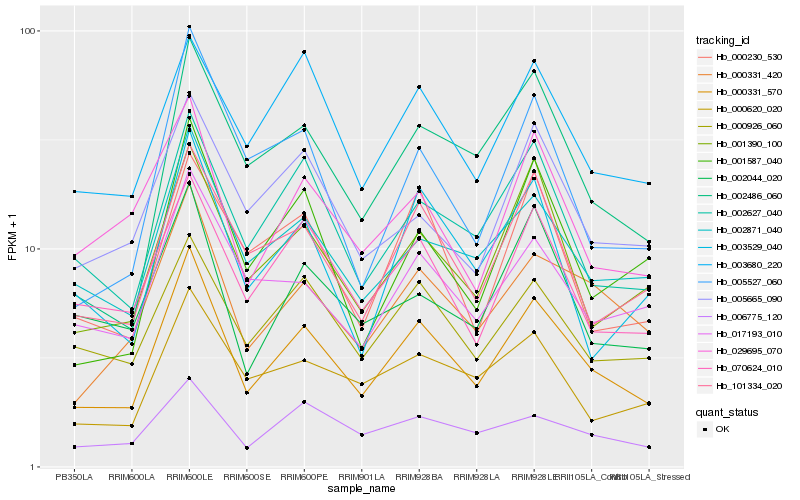
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_570 |
0.0 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase [Jatropha curcas] |
| 2 |
Hb_000331_420 |
0.0981039748 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 3 |
Hb_002871_040 |
0.1071971726 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 4 |
Hb_002627_040 |
0.1090424202 |
- |
- |
prolyl 4-hydroxylase alpha subunit, putative [Ricinus communis] |
| 5 |
Hb_101334_020 |
0.1102410986 |
- |
- |
PREDICTED: uncharacterized WD repeat-containing protein C2A9.03-like [Jatropha curcas] |
| 6 |
Hb_001390_100 |
0.1111631321 |
- |
- |
PREDICTED: LL-diaminopimelate aminotransferase, chloroplastic [Jatropha curcas] |
| 7 |
Hb_070624_010 |
0.1146404025 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 8 |
Hb_000926_060 |
0.1169412613 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_002486_060 |
0.1178561647 |
- |
- |
hypothetical protein JCGZ_01211 [Jatropha curcas] |
| 10 |
Hb_006775_120 |
0.1216317985 |
- |
- |
exonuclease-like protein [Oryza sativa Japonica Group] |
| 11 |
Hb_017193_010 |
0.1223959382 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 12 |
Hb_029695_070 |
0.1226876196 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_003680_220 |
0.1256806602 |
- |
- |
hypothetical protein B456_002G243700 [Gossypium raimondii] |
| 14 |
Hb_003529_040 |
0.1257713593 |
- |
- |
PREDICTED: ATP phosphoribosyltransferase 2, chloroplastic-like [Jatropha curcas] |
| 15 |
Hb_001587_040 |
0.1258898908 |
- |
- |
PREDICTED: soluble inorganic pyrophosphatase 6, chloroplastic [Jatropha curcas] |
| 16 |
Hb_002044_020 |
0.1273271438 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 17 |
Hb_000620_020 |
0.1274111396 |
- |
- |
PREDICTED: uncharacterized protein LOC105649917 [Jatropha curcas] |
| 18 |
Hb_005665_090 |
0.1279141932 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 19 |
Hb_005527_060 |
0.1296063482 |
- |
- |
malic enzyme, putative [Ricinus communis] |
| 20 |
Hb_000230_530 |
0.1309612583 |
- |
- |
conserved hypothetical protein [Ricinus communis] |