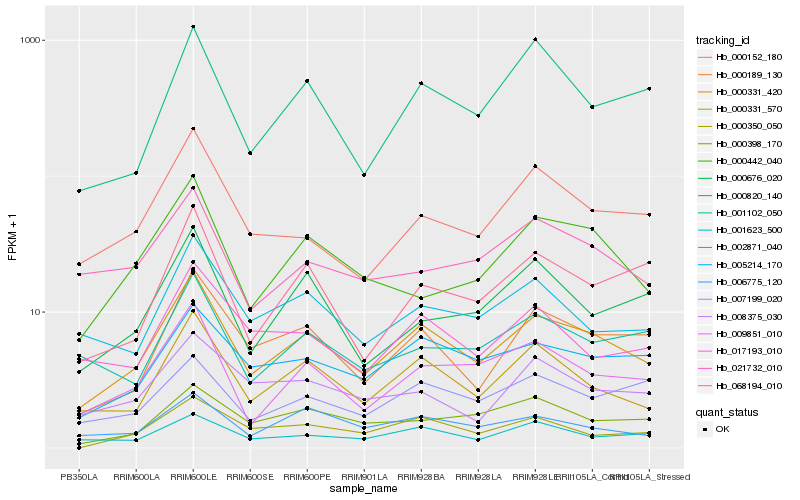
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000331_420 |
0.0 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 2 |
Hb_000331_570 |
0.0981039748 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase [Jatropha curcas] |
| 3 |
Hb_000189_130 |
0.1160462769 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATA, chloroplastic [Jatropha curcas] |
| 4 |
Hb_005214_170 |
0.1174907973 |
- |
- |
PREDICTED: uncharacterized protein LOC105636021 [Jatropha curcas] |
| 5 |
Hb_009851_010 |
0.1175792227 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_002871_040 |
0.1321104696 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 7 |
Hb_000152_180 |
0.1323962361 |
- |
- |
PREDICTED: CDGSH iron-sulfur domain-containing protein NEET [Jatropha curcas] |
| 8 |
Hb_006775_120 |
0.1331811083 |
- |
- |
exonuclease-like protein [Oryza sativa Japonica Group] |
| 9 |
Hb_001623_500 |
0.1371880664 |
- |
- |
PREDICTED: uncharacterized protein LOC105638487 [Jatropha curcas] |
| 10 |
Hb_017193_010 |
0.1389952767 |
- |
- |
PREDICTED: lactation elevated protein 1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_068194_010 |
0.1395738161 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 12 |
Hb_000820_140 |
0.1403763432 |
- |
- |
histone H4 [Zea mays] |
| 13 |
Hb_000442_040 |
0.1432890904 |
- |
- |
PREDICTED: biotin carboxyl carrier protein of acetyl-CoA carboxylase, chloroplastic [Jatropha curcas] |
| 14 |
Hb_007199_020 |
0.1460066167 |
- |
- |
hypothetical protein CICLE_v10028570mg [Citrus clementina] |
| 15 |
Hb_000350_050 |
0.1467434003 |
- |
- |
hypothetical protein JCGZ_07831 [Jatropha curcas] |
| 16 |
Hb_008375_030 |
0.1473967845 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 17 |
Hb_000676_020 |
0.1477054456 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 18 |
Hb_000398_170 |
0.1481875482 |
- |
- |
PREDICTED: tobamovirus multiplication protein 1-like [Jatropha curcas] |
| 19 |
Hb_021732_010 |
0.1502613282 |
- |
- |
biotin carboxylase [Vernicia fordii] |
| 20 |
Hb_001102_050 |
0.1505024627 |
- |
- |
PREDICTED: fatty-acid-binding protein 3 [Jatropha curcas] |