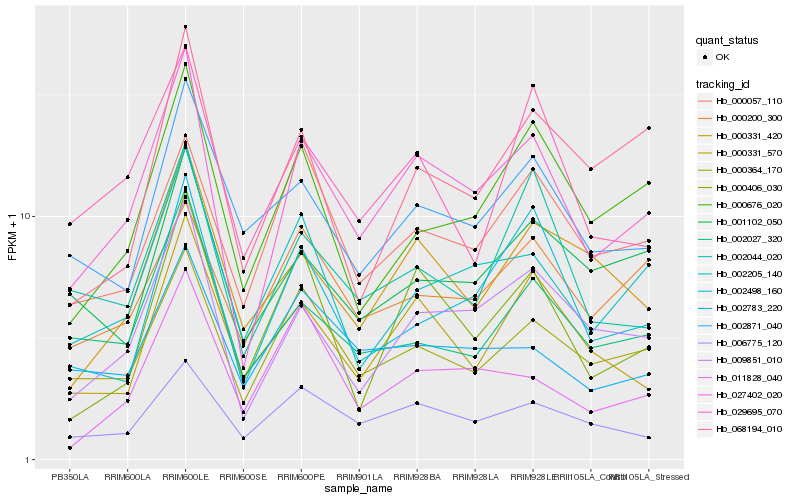
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027402_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 2 |
Hb_009851_010 |
0.1023029516 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic-like [Jatropha curcas] |
| 3 |
Hb_000364_170 |
0.1303396252 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 4 |
Hb_006775_120 |
0.1339001778 |
- |
- |
exonuclease-like protein [Oryza sativa Japonica Group] |
| 5 |
Hb_002205_140 |
0.1347399496 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 6 |
Hb_000406_030 |
0.1411353121 |
- |
- |
Bifunctional 3-dehydroquinate dehydratase/shikimate dehydrogenase [Morus notabilis] |
| 7 |
Hb_000331_570 |
0.1463294133 |
- |
- |
PREDICTED: DNA-(apurinic or apyrimidinic site) lyase [Jatropha curcas] |
| 8 |
Hb_000676_020 |
0.1464786707 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 9 |
Hb_002498_160 |
0.1476945685 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 10 |
Hb_002044_020 |
0.1494097236 |
- |
- |
glucose-1-phosphate denylyltransferase small subunit [Hevea brasiliensis] |
| 11 |
Hb_002027_320 |
0.1494851713 |
- |
- |
PREDICTED: RNA-binding protein BRN1 [Jatropha curcas] |
| 12 |
Hb_029695_070 |
0.1504215684 |
- |
- |
PREDICTED: nudix hydrolase 10-like isoform X1 [Jatropha curcas] |
| 13 |
Hb_000331_420 |
0.1510007524 |
- |
- |
PREDICTED: nucleoside diphosphate kinase 2, chloroplastic [Jatropha curcas] |
| 14 |
Hb_002783_220 |
0.1553623378 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 15 |
Hb_000057_110 |
0.1554874365 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 16 |
Hb_002871_040 |
0.1580487667 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 17 |
Hb_011828_040 |
0.1629110705 |
- |
- |
PREDICTED: uncharacterized protein LOC105642173 isoform X2 [Jatropha curcas] |
| 18 |
Hb_001102_050 |
0.1636359043 |
- |
- |
PREDICTED: fatty-acid-binding protein 3 [Jatropha curcas] |
| 19 |
Hb_000200_300 |
0.1643692436 |
- |
- |
PREDICTED: uncharacterized protein LOC105636926 [Jatropha curcas] |
| 20 |
Hb_068194_010 |
0.165015608 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |