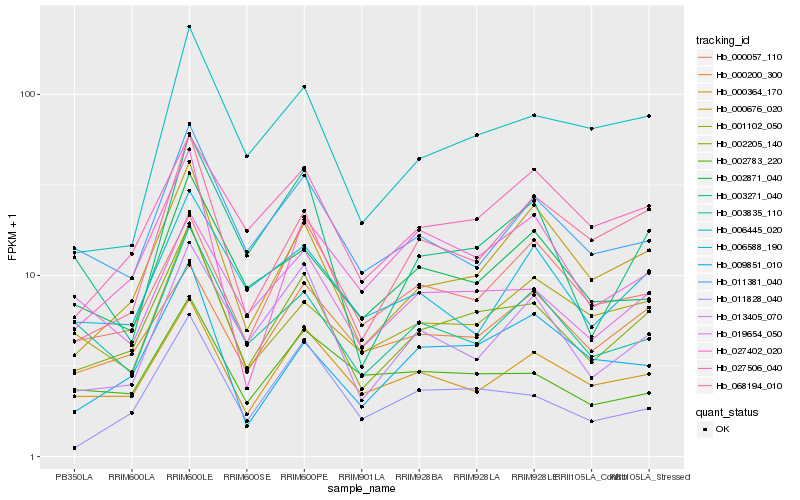
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002205_140 |
0.0 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 2 |
Hb_000676_020 |
0.1114633002 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 3 |
Hb_000364_170 |
0.1121316781 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 4 |
Hb_019654_050 |
0.1175804092 |
- |
- |
PREDICTED: prolyl endopeptidase [Jatropha curcas] |
| 5 |
Hb_011381_040 |
0.122283688 |
- |
- |
PREDICTED: uncharacterized protein At1g32220, chloroplastic [Jatropha curcas] |
| 6 |
Hb_013405_070 |
0.125369411 |
- |
- |
ferric-chelate reductase, putative [Ricinus communis] |
| 7 |
Hb_027506_040 |
0.1265797556 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_006445_020 |
0.1294211984 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |
| 9 |
Hb_011828_040 |
0.1311995176 |
- |
- |
PREDICTED: uncharacterized protein LOC105642173 isoform X2 [Jatropha curcas] |
| 10 |
Hb_000200_300 |
0.13278771 |
- |
- |
PREDICTED: uncharacterized protein LOC105636926 [Jatropha curcas] |
| 11 |
Hb_003835_110 |
0.1337053528 |
- |
- |
PREDICTED: nucleobase-ascorbate transporter 11 isoform X1 [Jatropha curcas] |
| 12 |
Hb_068194_010 |
0.1342035862 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 13 |
Hb_002871_040 |
0.1342948839 |
- |
- |
PREDICTED: enoyl-[acyl-carrier-protein] reductase [NADH], chloroplastic-like [Populus euphratica] |
| 14 |
Hb_027402_020 |
0.1347399496 |
- |
- |
PREDICTED: uncharacterized endoplasmic reticulum membrane protein C16E8.02 [Jatropha curcas] |
| 15 |
Hb_003271_040 |
0.1374878514 |
- |
- |
PREDICTED: SEC14 cytosolic factor [Jatropha curcas] |
| 16 |
Hb_000057_110 |
0.1385900374 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 17 |
Hb_009851_010 |
0.138944652 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic-like [Jatropha curcas] |
| 18 |
Hb_002783_220 |
0.139139573 |
- |
- |
PREDICTED: uncharacterized protein LOC105635342 isoform X1 [Jatropha curcas] |
| 19 |
Hb_001102_050 |
0.1407199615 |
- |
- |
PREDICTED: fatty-acid-binding protein 3 [Jatropha curcas] |
| 20 |
Hb_006588_190 |
0.1408105867 |
- |
- |
PREDICTED: ataxin-3 homolog [Jatropha curcas] |