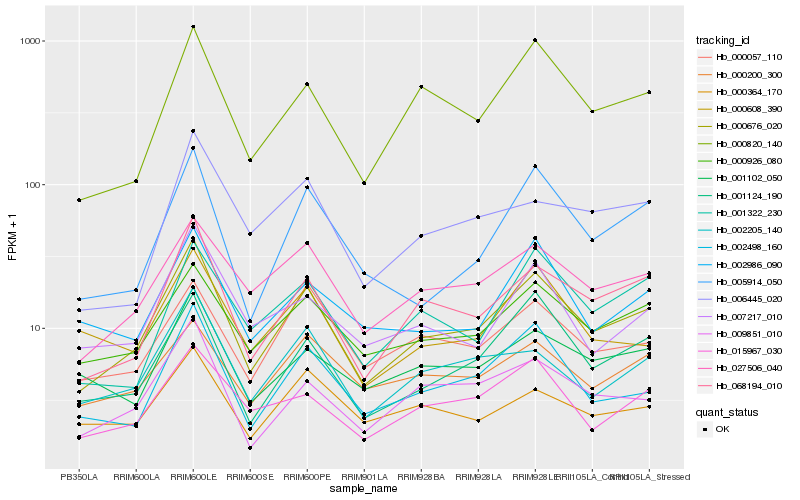
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000676_020 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 2 |
Hb_068194_010 |
0.0832281369 |
- |
- |
hypothetical protein POPTR_0010s23740g [Populus trichocarpa] |
| 3 |
Hb_027506_040 |
0.1066942705 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002498_160 |
0.1067116784 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 5 |
Hb_009851_010 |
0.1097882486 |
- |
- |
PREDICTED: beta-amylase 3, chloroplastic-like [Jatropha curcas] |
| 6 |
Hb_000820_140 |
0.1101443128 |
- |
- |
histone H4 [Zea mays] |
| 7 |
Hb_002205_140 |
0.1114633002 |
transcription factor |
TF Family: bZIP |
PREDICTED: transcription factor RF2b [Jatropha curcas] |
| 8 |
Hb_000926_080 |
0.1118200625 |
- |
- |
PREDICTED: paraspeckle component 1 [Jatropha curcas] |
| 9 |
Hb_001124_190 |
0.1129989907 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_015967_030 |
0.12092113 |
transcription factor |
TF Family: B3 |
PREDICTED: B3 domain-containing transcription repressor VAL1 isoform X1 [Jatropha curcas] |
| 11 |
Hb_005914_050 |
0.1211316122 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 12 |
Hb_007217_010 |
0.1272526465 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001102_050 |
0.1291703248 |
- |
- |
PREDICTED: fatty-acid-binding protein 3 [Jatropha curcas] |
| 14 |
Hb_006445_020 |
0.130618961 |
- |
- |
hypothetical protein L484_026741 [Morus notabilis] |
| 15 |
Hb_001322_230 |
0.1309329739 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 16 |
Hb_002986_090 |
0.1319798518 |
- |
- |
PREDICTED: methionine aminopeptidase 1D, chloroplastic/mitochondrial [Jatropha curcas] |
| 17 |
Hb_000200_300 |
0.133806041 |
- |
- |
PREDICTED: uncharacterized protein LOC105636926 [Jatropha curcas] |
| 18 |
Hb_000364_170 |
0.1340877361 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase CLF [Jatropha curcas] |
| 19 |
Hb_000057_110 |
0.1347543184 |
- |
- |
PREDICTED: kinesin-13A [Jatropha curcas] |
| 20 |
Hb_000608_390 |
0.1353461629 |
- |
- |
hypothetical protein B456_010G140700 [Gossypium raimondii] |