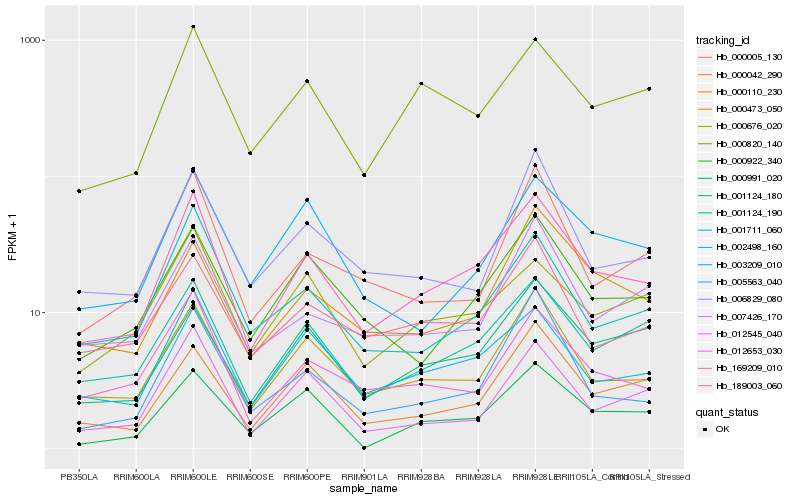
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_169209_010 |
0.0 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 2 |
Hb_002498_160 |
0.1004842358 |
- |
- |
PREDICTED: uncharacterized protein LOC105649315 [Jatropha curcas] |
| 3 |
Hb_000473_050 |
0.1041459712 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 4 |
Hb_001124_190 |
0.1079153422 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_000922_340 |
0.1259317346 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 6 |
Hb_012653_030 |
0.1267285495 |
- |
- |
PREDICTED: probable ribose-5-phosphate isomerase 4, chloroplastic isoform X2 [Jatropha curcas] |
| 7 |
Hb_000820_140 |
0.1290334333 |
- |
- |
histone H4 [Zea mays] |
| 8 |
Hb_000042_290 |
0.1349407578 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 9 |
Hb_001711_060 |
0.1349522037 |
- |
- |
PREDICTED: ferredoxin isoform X1 [Jatropha curcas] |
| 10 |
Hb_189003_060 |
0.1357233944 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_001124_180 |
0.1371808521 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 12 |
Hb_000676_020 |
0.1393671886 |
- |
- |
PREDICTED: uncharacterized protein LOC105645386 [Jatropha curcas] |
| 13 |
Hb_000991_020 |
0.1395757606 |
- |
- |
amino acid transporter, putative [Ricinus communis] |
| 14 |
Hb_012545_040 |
0.1400937504 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 15 |
Hb_003209_010 |
0.1423229492 |
- |
- |
hypothetical protein POPTR_0010s16880g [Populus trichocarpa] |
| 16 |
Hb_000110_230 |
0.1424790257 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_005563_040 |
0.1430939398 |
- |
- |
PREDICTED: uncharacterized protein LOC105630105 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000005_130 |
0.1432057321 |
- |
- |
hypothetical protein POPTR_0004s22660g [Populus trichocarpa] |
| 19 |
Hb_006829_080 |
0.1442702309 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 20 |
Hb_007426_170 |
0.1470195022 |
transcription factor |
TF Family: TCP |
- |