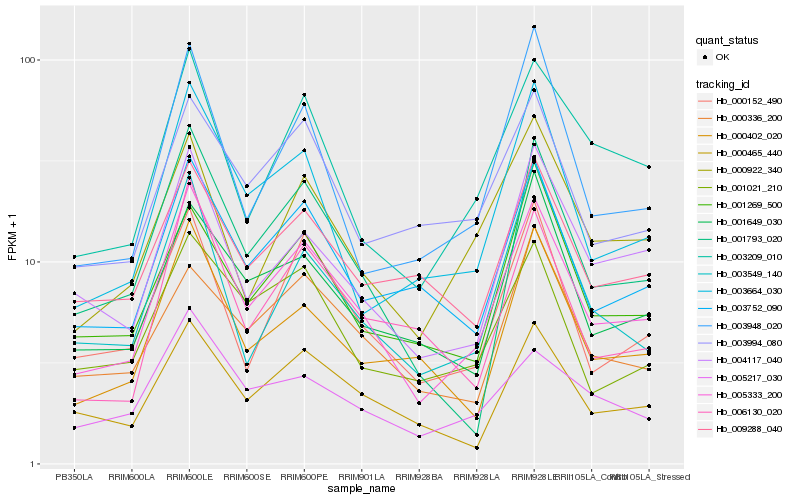
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005333_200 |
0.0 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_003752_090 |
0.112781434 |
- |
- |
chitinase, putative [Ricinus communis] |
| 3 |
Hb_003209_010 |
0.112933459 |
- |
- |
hypothetical protein POPTR_0010s16880g [Populus trichocarpa] |
| 4 |
Hb_000152_490 |
0.1144588107 |
- |
- |
PREDICTED: (6-4)DNA photolyase [Jatropha curcas] |
| 5 |
Hb_004117_040 |
0.116322248 |
- |
- |
PREDICTED: uncharacterized protein LOC105638287 [Jatropha curcas] |
| 6 |
Hb_000465_440 |
0.1169584029 |
- |
- |
PREDICTED: polycomb group protein EMBRYONIC FLOWER 2-like [Jatropha curcas] |
| 7 |
Hb_001793_020 |
0.1170954442 |
- |
- |
PREDICTED: uncharacterized protein LOC105628624 [Jatropha curcas] |
| 8 |
Hb_003664_030 |
0.1185237623 |
- |
- |
PREDICTED: mitochondrial carnitine/acylcarnitine carrier-like protein [Jatropha curcas] |
| 9 |
Hb_000922_340 |
0.1194535366 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 10 |
Hb_000402_020 |
0.1266678279 |
- |
- |
PREDICTED: 2-phytyl-1,4-beta-naphthoquinone methyltransferase, chloroplastic isoform X1 [Jatropha curcas] |
| 11 |
Hb_001021_210 |
0.1275630382 |
transcription factor |
TF Family: mTERF |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_001649_030 |
0.1278903296 |
- |
- |
PREDICTED: peptide deformylase 1B, chloroplastic [Jatropha curcas] |
| 13 |
Hb_005217_030 |
0.1281266996 |
- |
- |
PREDICTED: probable transcriptional regulator SLK3 [Jatropha curcas] |
| 14 |
Hb_003549_140 |
0.1291914694 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 15 |
Hb_003994_080 |
0.1295399756 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 16 |
Hb_006130_020 |
0.1302525689 |
- |
- |
S-adenosylmethionine-dependent methyltransferase, putative [Ricinus communis] |
| 17 |
Hb_001269_500 |
0.1326304405 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 18 |
Hb_009288_040 |
0.133062186 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 19 |
Hb_003948_020 |
0.1337046038 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 20 |
Hb_000336_200 |
0.1345332531 |
- |
- |
PREDICTED: UPF0481 protein At3g47200-like [Jatropha curcas] |