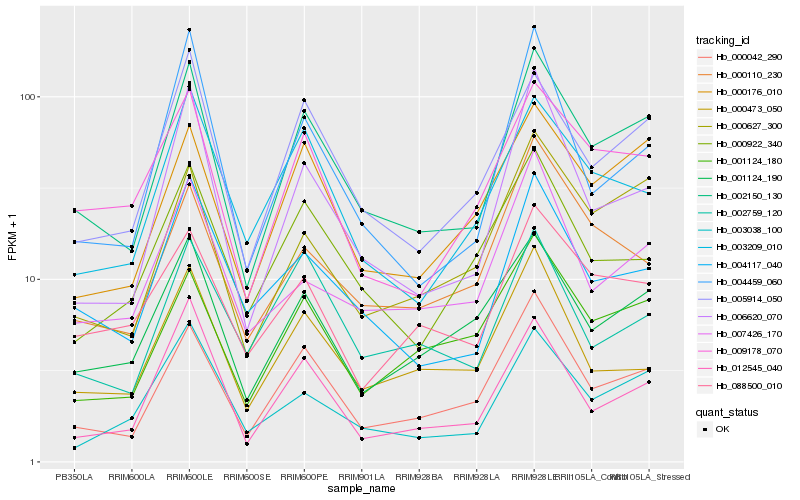
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_002150_130 |
0.0 |
- |
- |
50S ribosomal protein L18, chloroplast precursor, putative [Ricinus communis] |
| 2 |
Hb_000042_290 |
0.0911524318 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 3 |
Hb_005914_050 |
0.0997677045 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 4 |
Hb_000176_010 |
0.1108814396 |
- |
- |
PREDICTED: 50S ribosomal protein L35, chloroplastic [Jatropha curcas] |
| 5 |
Hb_000627_300 |
0.1151772753 |
- |
- |
PREDICTED: uncharacterized protein LOC105632672 [Jatropha curcas] |
| 6 |
Hb_009178_070 |
0.1152560337 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 8, chloroplastic [Jatropha curcas] |
| 7 |
Hb_004117_040 |
0.1169367168 |
- |
- |
PREDICTED: uncharacterized protein LOC105638287 [Jatropha curcas] |
| 8 |
Hb_006620_070 |
0.1209431124 |
- |
- |
chloroplast 30S ribosomal protein S13 [Hevea brasiliensis] |
| 9 |
Hb_001124_190 |
0.1218646511 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_001124_180 |
0.1255147828 |
- |
- |
PREDICTED: uncharacterized protein LOC105171432 [Sesamum indicum] |
| 11 |
Hb_003038_100 |
0.1268858447 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 12 |
Hb_003209_010 |
0.1311359315 |
- |
- |
hypothetical protein POPTR_0010s16880g [Populus trichocarpa] |
| 13 |
Hb_012545_040 |
0.1359614074 |
- |
- |
PREDICTED: uroporphyrinogen-III synthase, chloroplastic isoform X1 [Jatropha curcas] |
| 14 |
Hb_088500_010 |
0.1362648855 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At4g39970 [Jatropha curcas] |
| 15 |
Hb_000110_230 |
0.1397942025 |
- |
- |
Plastid-specific 30S ribosomal protein 3, chloroplast precursor, putative [Ricinus communis] |
| 16 |
Hb_007426_170 |
0.1405002506 |
transcription factor |
TF Family: TCP |
- |
| 17 |
Hb_000922_340 |
0.1417135652 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 18 |
Hb_000473_050 |
0.143047696 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 19 |
Hb_002759_120 |
0.1430971321 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_004459_060 |
0.1437027919 |
- |
- |
50S ribosomal protein L34, chloroplast precursor, putative [Ricinus communis] |