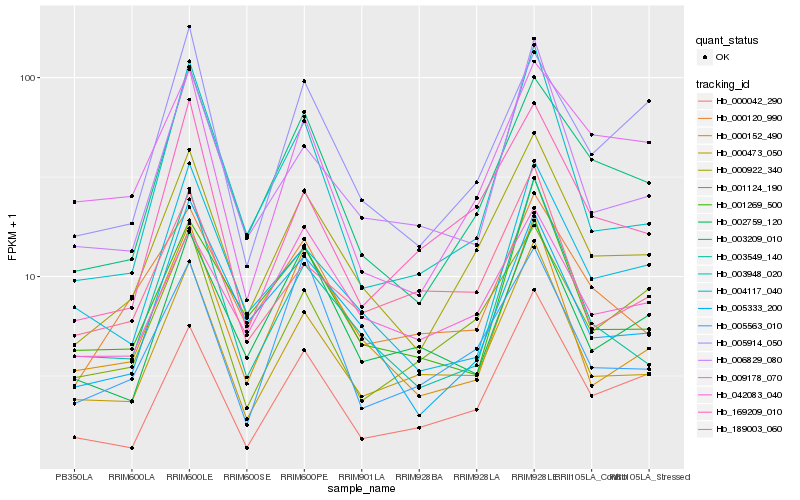
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000922_340 |
0.0 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 2 |
Hb_003209_010 |
0.0900799651 |
- |
- |
hypothetical protein POPTR_0010s16880g [Populus trichocarpa] |
| 3 |
Hb_000473_050 |
0.1095594919 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 4 |
Hb_000042_290 |
0.1176529336 |
- |
- |
PREDICTED: uncharacterized protein LOC105632818 isoform X2 [Jatropha curcas] |
| 5 |
Hb_042083_040 |
0.117938215 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_005333_200 |
0.1194535366 |
- |
- |
PREDICTED: beta carbonic anhydrase 5, chloroplastic-like [Jatropha curcas] |
| 7 |
Hb_000120_990 |
0.1208075407 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_001124_190 |
0.1248111407 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 9 |
Hb_005914_050 |
0.125229523 |
- |
- |
PREDICTED: 50S ribosomal protein L28, chloroplastic [Jatropha curcas] |
| 10 |
Hb_169209_010 |
0.1259317346 |
- |
- |
PREDICTED: porphobilinogen deaminase, chloroplastic [Jatropha curcas] |
| 11 |
Hb_006829_080 |
0.1294511097 |
- |
- |
PREDICTED: protein LHCP TRANSLOCATION DEFECT [Jatropha curcas] |
| 12 |
Hb_000152_490 |
0.1324503222 |
- |
- |
PREDICTED: (6-4)DNA photolyase [Jatropha curcas] |
| 13 |
Hb_005563_010 |
0.132932008 |
- |
- |
PREDICTED: kinesin-II 85 kDa subunit isoform X1 [Jatropha curcas] |
| 14 |
Hb_003948_020 |
0.1346838836 |
- |
- |
PREDICTED: transmembrane protein 41B [Jatropha curcas] |
| 15 |
Hb_009178_070 |
0.1355663873 |
- |
- |
PREDICTED: probable plastid-lipid-associated protein 8, chloroplastic [Jatropha curcas] |
| 16 |
Hb_003549_140 |
0.1361067512 |
- |
- |
Thylakoid lumenal 21.5 kDa protein, chloroplast precursor, putative [Ricinus communis] |
| 17 |
Hb_001269_500 |
0.1368678859 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 18 |
Hb_002759_120 |
0.1369486665 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_004117_040 |
0.1370604793 |
- |
- |
PREDICTED: uncharacterized protein LOC105638287 [Jatropha curcas] |
| 20 |
Hb_189003_060 |
0.1371871344 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |