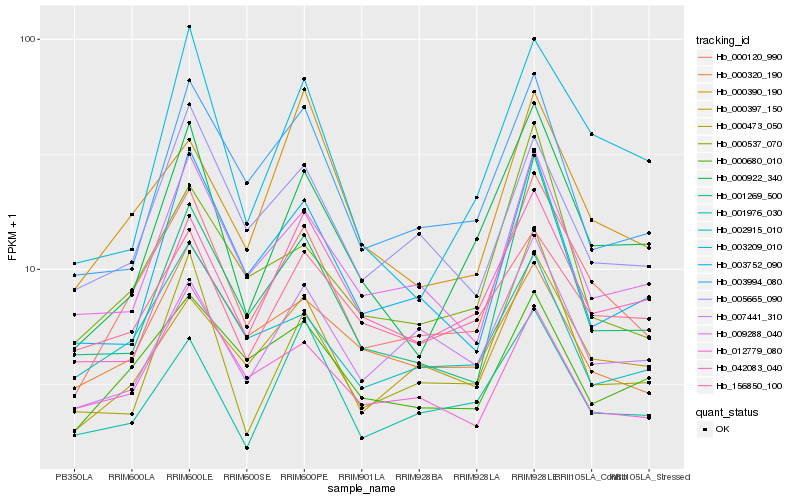
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000120_990 |
0.0 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic-like [Jatropha curcas] |
| 2 |
Hb_000397_150 |
0.1039278434 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 3 |
Hb_009288_040 |
0.1178718874 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 4 |
Hb_000922_340 |
0.1208075407 |
- |
- |
PREDICTED: uncharacterized protein LOC105640368 [Jatropha curcas] |
| 5 |
Hb_000680_010 |
0.1220430082 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Populus euphratica] |
| 6 |
Hb_001269_500 |
0.1263784829 |
- |
- |
PREDICTED: protein LOW PSII ACCUMULATION 3, chloroplastic-like isoform X2 [Populus euphratica] |
| 7 |
Hb_003994_080 |
0.1267733814 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 8 |
Hb_005665_090 |
0.1269646797 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 9 |
Hb_042083_040 |
0.128313331 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 10 |
Hb_007441_310 |
0.1306550481 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_001976_030 |
0.1314408249 |
- |
- |
PREDICTED: protein NETWORKED 2D-like isoform X2 [Jatropha curcas] |
| 12 |
Hb_000537_070 |
0.1325476433 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At5g25630 [Jatropha curcas] |
| 13 |
Hb_156850_100 |
0.1326966667 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 14 |
Hb_002915_010 |
0.1330140369 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 15 |
Hb_000390_190 |
0.1331740476 |
- |
- |
ATP binding protein, putative [Ricinus communis] |
| 16 |
Hb_000320_190 |
0.1333967995 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 17 |
Hb_000473_050 |
0.1342335617 |
- |
- |
chromatin regulatory protein sir2, putative [Ricinus communis] |
| 18 |
Hb_012779_080 |
0.1350133061 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 19 |
Hb_003752_090 |
0.1354780429 |
- |
- |
chitinase, putative [Ricinus communis] |
| 20 |
Hb_003209_010 |
0.1355163173 |
- |
- |
hypothetical protein POPTR_0010s16880g [Populus trichocarpa] |