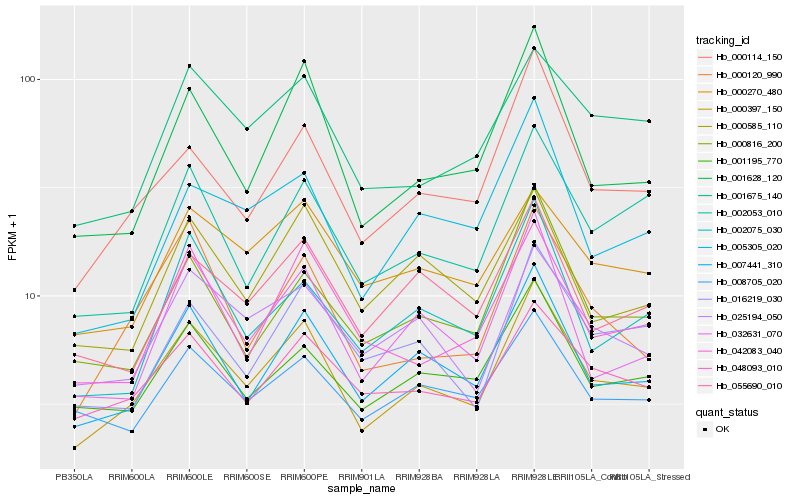
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000397_150 |
0.0 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |
| 2 |
Hb_007441_310 |
0.072428355 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000114_150 |
0.0852243008 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 4 |
Hb_001628_120 |
0.0911428273 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 5 |
Hb_048093_010 |
0.0970763313 |
- |
- |
PREDICTED: calcium uniporter protein 6, mitochondrial-like [Jatropha curcas] |
| 6 |
Hb_001195_770 |
0.0978172389 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 7 |
Hb_032631_070 |
0.0986777056 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 8 |
Hb_025194_050 |
0.1010219083 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 9 |
Hb_055690_010 |
0.1021583122 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 10 |
Hb_000816_200 |
0.1021997464 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 11 |
Hb_001675_140 |
0.1028160817 |
- |
- |
hypothetical protein L484_026216 [Morus notabilis] |
| 12 |
Hb_000120_990 |
0.1039278434 |
- |
- |
PREDICTED: MATE efflux family protein 3, chloroplastic-like [Jatropha curcas] |
| 13 |
Hb_002053_010 |
0.1040115565 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_042083_040 |
0.1048407338 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 15 |
Hb_000270_480 |
0.1051264163 |
- |
- |
Mannose-1-phosphate guanyltransferase alpha [Gossypium arboreum] |
| 16 |
Hb_000585_110 |
0.1065913758 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 17 |
Hb_008705_020 |
0.1077369752 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 18 |
Hb_016219_030 |
0.1081520567 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 19 |
Hb_002075_030 |
0.1082848349 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 20 |
Hb_005305_020 |
0.109118636 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |