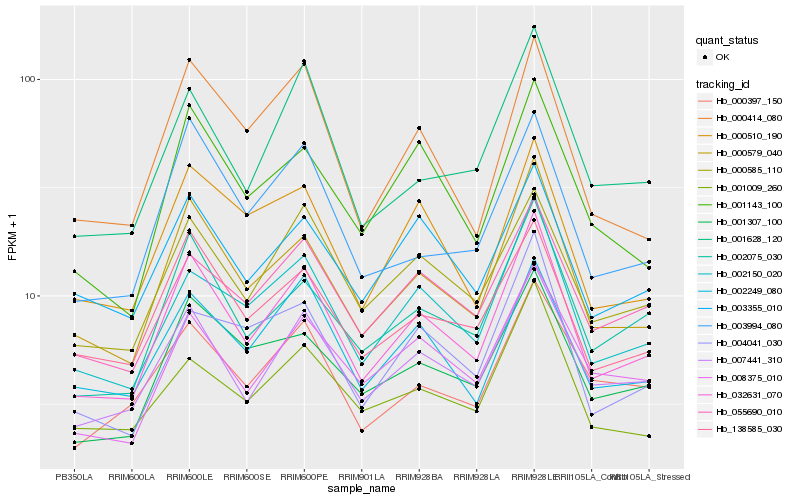
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_032631_070 |
0.0 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 2 |
Hb_000579_040 |
0.0484034532 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 3 |
Hb_007441_310 |
0.0681284984 |
- |
- |
PREDICTED: protein TRIGALACTOSYLDIACYLGLYCEROL 4, chloroplastic [Jatropha curcas] |
| 4 |
Hb_002075_030 |
0.0741006873 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 5 |
Hb_002150_020 |
0.0748567827 |
- |
- |
PREDICTED: translation initiation factor IF-2, mitochondrial isoform X1 [Jatropha curcas] |
| 6 |
Hb_138585_030 |
0.0865701383 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 7 |
Hb_000585_110 |
0.0871062752 |
- |
- |
PREDICTED: probable protein phosphatase 2C 66 [Jatropha curcas] |
| 8 |
Hb_001307_100 |
0.0887306754 |
- |
- |
calmodulin binding protein, putative [Ricinus communis] |
| 9 |
Hb_001628_120 |
0.0911460588 |
- |
- |
putative ascorbate peroxidase, partial [Taraxacum brevicorniculatum] |
| 10 |
Hb_008375_010 |
0.0915565699 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X3 [Pyrus x bretschneideri] |
| 11 |
Hb_002249_080 |
0.091893133 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 12 |
Hb_055690_010 |
0.0929057754 |
- |
- |
PREDICTED: pheophytinase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_001143_100 |
0.0947077164 |
- |
- |
PREDICTED: delta-aminolevulinic acid dehydratase, chloroplastic [Populus euphratica] |
| 14 |
Hb_003994_080 |
0.0950583546 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |
| 15 |
Hb_004041_030 |
0.0964949565 |
- |
- |
PREDICTED: mitochondrial substrate carrier family protein C-like isoform X1 [Populus euphratica] |
| 16 |
Hb_000510_190 |
0.0967151098 |
- |
- |
glutathione reductase [Hevea brasiliensis] |
| 17 |
Hb_001009_260 |
0.0978656198 |
- |
- |
PREDICTED: uncharacterized aarF domain-containing protein kinase 1 isoform X1 [Jatropha curcas] |
| 18 |
Hb_000414_080 |
0.0981490703 |
- |
- |
PREDICTED: triosephosphate isomerase, chloroplastic [Jatropha curcas] |
| 19 |
Hb_003355_010 |
0.0985534183 |
- |
- |
Heat shock 70 kDa protein, putative [Ricinus communis] |
| 20 |
Hb_000397_150 |
0.0986777056 |
- |
- |
PREDICTED: probable flavin-containing monooxygenase 1 [Jatropha curcas] |