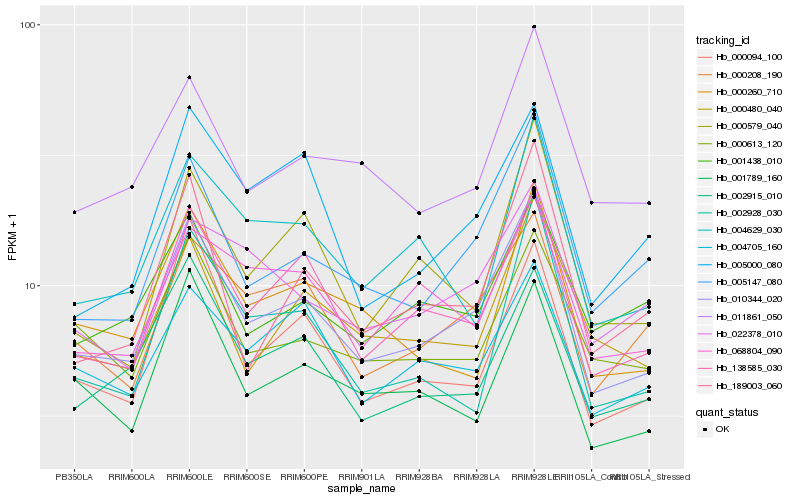
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_010344_020 |
0.0 |
- |
- |
PREDICTED: pentatricopeptide repeat-containing protein At1g79490, mitochondrial [Jatropha curcas] |
| 2 |
Hb_138585_030 |
0.0753677824 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 3 |
Hb_005147_080 |
0.084490401 |
- |
- |
PREDICTED: prolycopene isomerase, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_011861_050 |
0.0934382352 |
- |
- |
PREDICTED: COBW domain-containing protein 1 [Jatropha curcas] |
| 5 |
Hb_002915_010 |
0.0935439921 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 6 |
Hb_000208_190 |
0.0941242986 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 7 |
Hb_000094_100 |
0.0965471487 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 8 |
Hb_001789_160 |
0.1005195634 |
- |
- |
PREDICTED: putative transporter arsB [Jatropha curcas] |
| 9 |
Hb_000480_040 |
0.1005904738 |
- |
- |
PREDICTED: L-Ala-D/L-amino acid epimerase isoform X2 [Jatropha curcas] |
| 10 |
Hb_000260_710 |
0.1027467889 |
- |
- |
PREDICTED: uncharacterized protein LOC105649017 isoform X1 [Jatropha curcas] |
| 11 |
Hb_189003_060 |
0.1032501509 |
- |
- |
PREDICTED: uroporphyrinogen decarboxylase 1, chloroplastic isoform X1 [Jatropha curcas] |
| 12 |
Hb_000613_120 |
0.1044591275 |
transcription factor |
TF Family: SET |
PREDICTED: ribulose-1,5 bisphosphate carboxylase/oxygenase large subunit N-methyltransferase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_068804_090 |
0.1053154911 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 14 |
Hb_004705_160 |
0.1053846424 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 15 |
Hb_000579_040 |
0.1063135828 |
- |
- |
PREDICTED: putative GTP-binding protein 6 [Jatropha curcas] |
| 16 |
Hb_004629_030 |
0.1066326495 |
- |
- |
ABC transporter family protein [Hevea brasiliensis] |
| 17 |
Hb_002928_030 |
0.1088983229 |
- |
- |
hypothetical protein CICLE_v10001788mg [Citrus clementina] |
| 18 |
Hb_001438_010 |
0.1105931314 |
- |
- |
PREDICTED: uncharacterized protein LOC105639111 isoform X1 [Jatropha curcas] |
| 19 |
Hb_005000_080 |
0.1113182209 |
- |
- |
PREDICTED: thioredoxin-like 2, chloroplastic [Populus euphratica] |
| 20 |
Hb_022378_010 |
0.1121903544 |
- |
- |
PREDICTED: uncharacterized protein LOC105634715 isoform X1 [Jatropha curcas] |