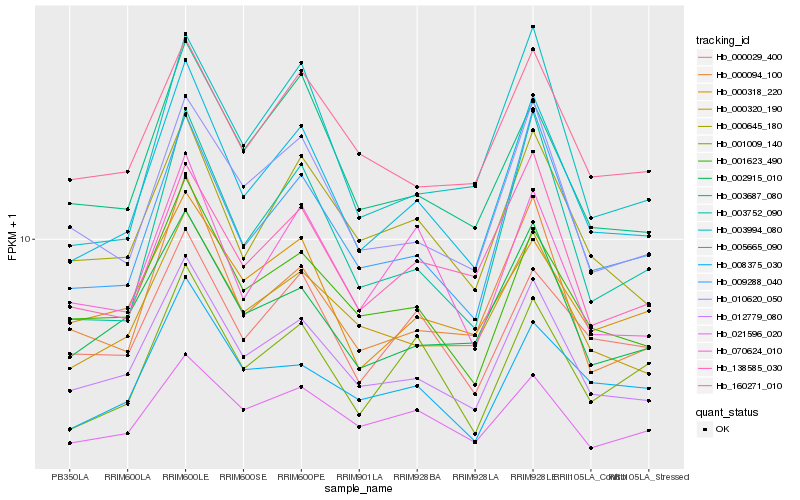
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_005665_090 |
0.0 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 2 |
Hb_012779_080 |
0.0518747735 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 3 |
Hb_009288_040 |
0.0717073985 |
- |
- |
PREDICTED: uncharacterized protein LOC105648677 [Jatropha curcas] |
| 4 |
Hb_001623_490 |
0.0792802075 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 5 |
Hb_000320_190 |
0.082399226 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 6 |
Hb_000645_180 |
0.0831025011 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 7 |
Hb_010620_050 |
0.0841803386 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 8 |
Hb_003752_090 |
0.0858644262 |
- |
- |
chitinase, putative [Ricinus communis] |
| 9 |
Hb_000029_400 |
0.0870739241 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |
| 10 |
Hb_000094_100 |
0.0882513637 |
- |
- |
PREDICTED: probable methyltransferase PMT28 [Jatropha curcas] |
| 11 |
Hb_003687_080 |
0.0887041102 |
- |
- |
PREDICTED: casein kinase I-like [Jatropha curcas] |
| 12 |
Hb_002915_010 |
0.0900963981 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 13 |
Hb_001009_140 |
0.0940004327 |
- |
- |
hypothetical protein JCGZ_11337 [Jatropha curcas] |
| 14 |
Hb_070624_010 |
0.0957001026 |
- |
- |
PREDICTED: reticulocalbin-2 [Jatropha curcas] |
| 15 |
Hb_138585_030 |
0.0970310516 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 16 |
Hb_008375_030 |
0.0970855701 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 4 isoform X1 [Populus euphratica] |
| 17 |
Hb_160271_010 |
0.0973560555 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 18 |
Hb_000318_220 |
0.098172225 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_021596_020 |
0.0984970187 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 20 |
Hb_003994_080 |
0.0984977976 |
- |
- |
PREDICTED: uncharacterized protein LOC105634384 [Jatropha curcas] |