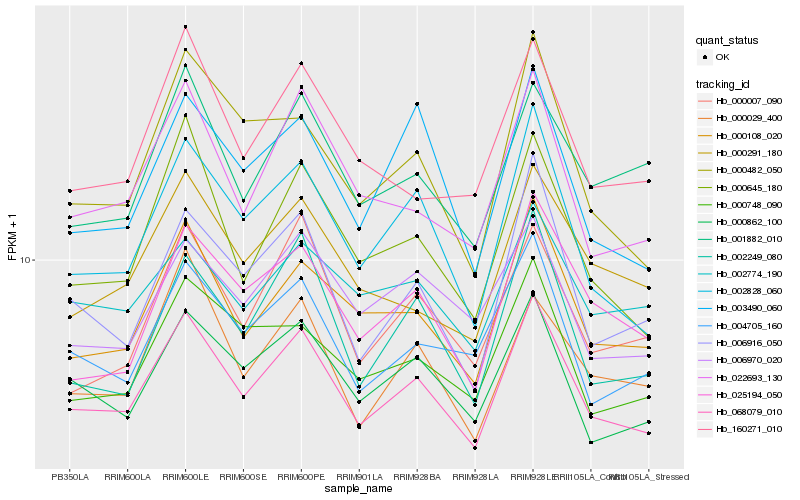
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_068079_010 |
0.0 |
- |
- |
- |
| 2 |
Hb_002828_060 |
0.0633482076 |
- |
- |
PREDICTED: probable phytol kinase 3, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000108_020 |
0.0656104257 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 4 |
Hb_000862_100 |
0.0687399724 |
- |
- |
PREDICTED: plastid division protein CDP1, chloroplastic [Jatropha curcas] |
| 5 |
Hb_022693_130 |
0.0739382662 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 6 |
Hb_000645_180 |
0.0746748231 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 7 |
Hb_004705_160 |
0.0772544669 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 8 |
Hb_006916_050 |
0.0772671143 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 9 |
Hb_002249_080 |
0.0823403701 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 10 |
Hb_025194_050 |
0.0828562716 |
- |
- |
PREDICTED: uncharacterized protein LOC105644851 isoform X1 [Jatropha curcas] |
| 11 |
Hb_000748_090 |
0.0853400667 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 12 |
Hb_000482_050 |
0.0862018951 |
- |
- |
PREDICTED: UDP-sulfoquinovose synthase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_003490_060 |
0.0863170775 |
- |
- |
PREDICTED: probable cyclic nucleotide-gated ion channel 20, chloroplastic isoform X2 [Sesamum indicum] |
| 14 |
Hb_006970_020 |
0.0867037743 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 15 |
Hb_000291_180 |
0.0867690092 |
- |
- |
PREDICTED: meiotically up-regulated gene 185 protein [Jatropha curcas] |
| 16 |
Hb_000007_090 |
0.0876345817 |
- |
- |
PREDICTED: LAG1 longevity assurance homolog 3 [Jatropha curcas] |
| 17 |
Hb_001882_010 |
0.0878751223 |
- |
- |
- |
| 18 |
Hb_160271_010 |
0.0881487844 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 19 |
Hb_002774_190 |
0.0885935995 |
- |
- |
Endoplasmic reticulum-Golgi intermediate compartment protein, putative [Ricinus communis] |
| 20 |
Hb_000029_400 |
0.0889134694 |
- |
- |
PREDICTED: probable 1-acyl-sn-glycerol-3-phosphate acyltransferase 5 isoform X1 [Jatropha curcas] |