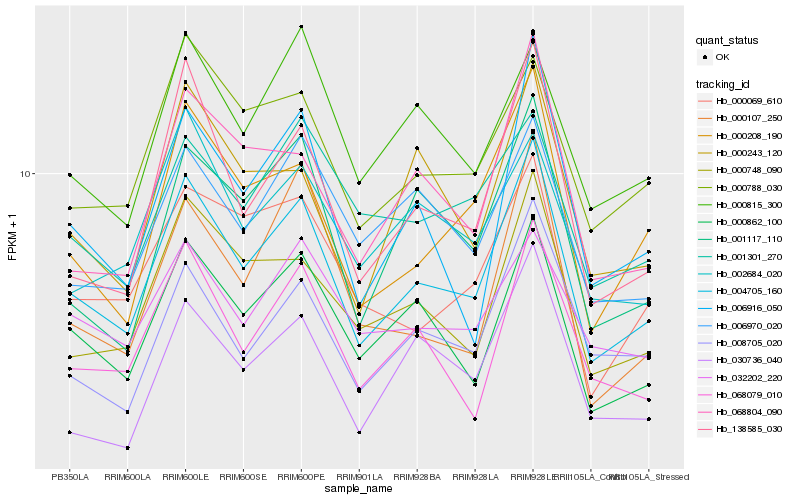
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_004705_160 |
0.0 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 2 |
Hb_000862_100 |
0.0645792383 |
- |
- |
PREDICTED: plastid division protein CDP1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_138585_030 |
0.0698495399 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 4 |
Hb_000208_190 |
0.0753091965 |
- |
- |
PREDICTED: uncharacterized protein LOC105648580 [Jatropha curcas] |
| 5 |
Hb_000748_090 |
0.0758076964 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |
| 6 |
Hb_068079_010 |
0.0772544669 |
- |
- |
- |
| 7 |
Hb_000788_030 |
0.0776994728 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 8 |
Hb_008705_020 |
0.0784603488 |
- |
- |
PREDICTED: lisH domain and HEAT repeat-containing protein KIAA1468 homolog isoform X1 [Jatropha curcas] |
| 9 |
Hb_068804_090 |
0.081473867 |
- |
- |
PREDICTED: probable cytosolic oligopeptidase A [Jatropha curcas] |
| 10 |
Hb_001117_110 |
0.0820467625 |
- |
- |
PREDICTED: dynamin-2A [Jatropha curcas] |
| 11 |
Hb_000815_300 |
0.0830871674 |
- |
- |
Transmembrane emp24 domain-containing protein 10 precursor, putative [Ricinus communis] |
| 12 |
Hb_030736_040 |
0.0831897924 |
- |
- |
PREDICTED: uncharacterized protein LOC105638926 isoform X1 [Jatropha curcas] |
| 13 |
Hb_006970_020 |
0.0847962917 |
- |
- |
PREDICTED: DNA topoisomerase 6 subunit B [Jatropha curcas] |
| 14 |
Hb_000069_610 |
0.0869314928 |
- |
- |
PREDICTED: protein SUPPRESSOR OF PHYA-105 1 [Jatropha curcas] |
| 15 |
Hb_002684_020 |
0.0871705785 |
- |
- |
ATP-dependent clp protease ATP-binding subunit clpx, putative [Ricinus communis] |
| 16 |
Hb_000243_120 |
0.0874116019 |
- |
- |
PREDICTED: MORC family CW-type zinc finger protein 3-like isoform X2 [Jatropha curcas] |
| 17 |
Hb_006916_050 |
0.0899352361 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 18 |
Hb_032202_220 |
0.0903691974 |
- |
- |
ring finger protein, putative [Ricinus communis] |
| 19 |
Hb_001301_270 |
0.0910078068 |
- |
- |
PREDICTED: heat shock 70 kDa protein 16 [Jatropha curcas] |
| 20 |
Hb_000107_250 |
0.0923160162 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |