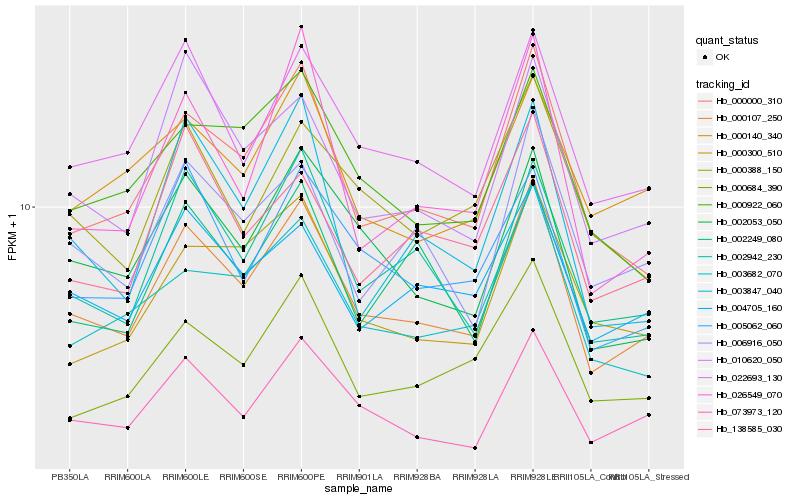
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000107_250 |
0.0 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 2 |
Hb_000000_310 |
0.0717016784 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 3 |
Hb_073973_120 |
0.0805621509 |
- |
- |
zinc finger protein, putative [Ricinus communis] |
| 4 |
Hb_022693_130 |
0.0807950686 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 5 |
Hb_002053_050 |
0.084286063 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 6 |
Hb_003682_070 |
0.0896592683 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 7 |
Hb_004705_160 |
0.0923160162 |
- |
- |
PREDICTED: protein MEI2-like 4 [Jatropha curcas] |
| 8 |
Hb_010620_050 |
0.0925684085 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 9 |
Hb_005062_060 |
0.0927318997 |
- |
- |
PREDICTED: uncharacterized protein At4g06598-like [Jatropha curcas] |
| 10 |
Hb_006916_050 |
0.095846874 |
- |
- |
PREDICTED: ultraviolet-B receptor UVR8 [Jatropha curcas] |
| 11 |
Hb_000684_390 |
0.0986834781 |
- |
- |
PREDICTED: mediator of RNA polymerase II transcription subunit 4 [Jatropha curcas] |
| 12 |
Hb_002249_080 |
0.099028942 |
- |
- |
lysosomal alpha-mannosidase, putative [Ricinus communis] |
| 13 |
Hb_000922_060 |
0.1006946335 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 14 |
Hb_026549_070 |
0.1026659595 |
- |
- |
PREDICTED: UDP-glucuronic acid decarboxylase 1 [Jatropha curcas] |
| 15 |
Hb_000300_510 |
0.1031174795 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 16 |
Hb_000388_150 |
0.1048322845 |
- |
- |
hypothetical protein JCGZ_21651 [Jatropha curcas] |
| 17 |
Hb_000140_340 |
0.1053634365 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 18 |
Hb_002942_230 |
0.1062289331 |
- |
- |
PREDICTED: long chain base biosynthesis protein 2a [Populus euphratica] |
| 19 |
Hb_003847_040 |
0.1064557452 |
- |
- |
adenosine diphosphatase, putative [Ricinus communis] |
| 20 |
Hb_138585_030 |
0.1074693761 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |