| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
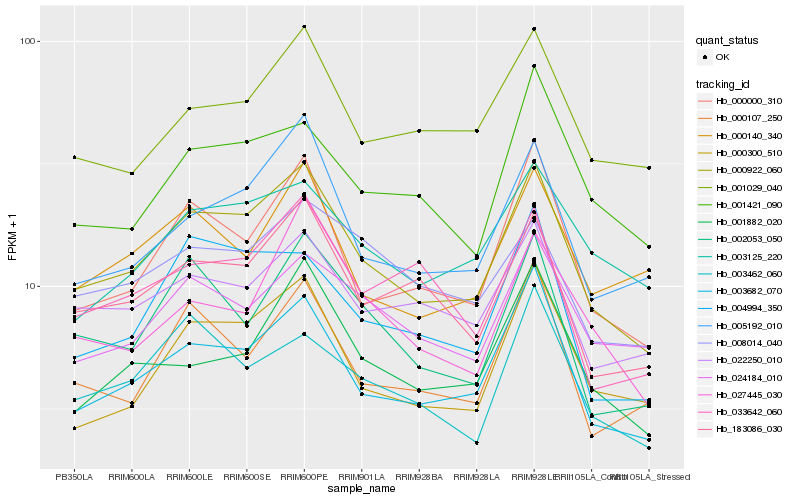
Hb_000922_060 |
0.0 |
- |
- |
kinesin light chain, putative [Ricinus communis] |
| 2 |
Hb_003682_070 |
0.0671361431 |
- |
- |
PREDICTED: uncharacterized protein LOC105633817 isoform X1 [Jatropha curcas] |
| 3 |
Hb_000000_310 |
0.0832653569 |
- |
- |
metalloendopeptidase, putative [Ricinus communis] |
| 4 |
Hb_005192_010 |
0.0859832059 |
- |
- |
PREDICTED: U-box domain-containing protein 4 [Jatropha curcas] |
| 5 |
Hb_183086_030 |
0.0877084965 |
- |
- |
GTPase-activating protein GYP7 [Gossypium arboreum] |
| 6 |
Hb_000300_510 |
0.0885941183 |
- |
- |
PREDICTED: uncharacterized protein LOC105632605 [Jatropha curcas] |
| 7 |
Hb_022250_010 |
0.0949975432 |
- |
- |
PREDICTED: probable beta-1,4-xylosyltransferase IRX10L [Jatropha curcas] |
| 8 |
Hb_002053_050 |
0.096175891 |
- |
- |
Kinesin heavy chain, putative [Ricinus communis] |
| 9 |
Hb_001029_040 |
0.1005885781 |
- |
- |
PREDICTED: uncharacterized protein LOC105641058 [Jatropha curcas] |
| 10 |
Hb_001882_020 |
0.100589039 |
- |
- |
PREDICTED: uncharacterized protein LOC105628417 [Jatropha curcas] |
| 11 |
Hb_004994_350 |
0.1006072333 |
- |
- |
PREDICTED: UPF0187 protein At3g61320, chloroplastic [Jatropha curcas] |
| 12 |
Hb_000107_250 |
0.1006946335 |
- |
- |
voltage-gated clc-type chloride channel, putative [Ricinus communis] |
| 13 |
Hb_024184_010 |
0.1018404361 |
- |
- |
PREDICTED: clustered mitochondria protein-like isoform X1 [Citrus sinensis] |
| 14 |
Hb_001421_090 |
0.1022892111 |
- |
- |
hypothetical protein RCOM_1278080 [Ricinus communis] |
| 15 |
Hb_003125_220 |
0.1037704466 |
- |
- |
PREDICTED: CBS domain-containing protein CBSCBSPB1 [Jatropha curcas] |
| 16 |
Hb_003462_060 |
0.104578292 |
- |
- |
PREDICTED: ABC transporter G family member 24-like [Jatropha curcas] |
| 17 |
Hb_008014_040 |
0.1050318331 |
- |
- |
PREDICTED: uncharacterized protein LOC105649721 [Jatropha curcas] |
| 18 |
Hb_033642_060 |
0.1050499613 |
- |
- |
PREDICTED: protein CHUP1, chloroplastic [Jatropha curcas] |
| 19 |
Hb_000140_340 |
0.1099114912 |
- |
- |
PREDICTED: protein ABIL1 [Jatropha curcas] |
| 20 |
Hb_027445_030 |
0.1100365671 |
- |
- |
F-box/LRR-repeat protein, putative [Ricinus communis] |