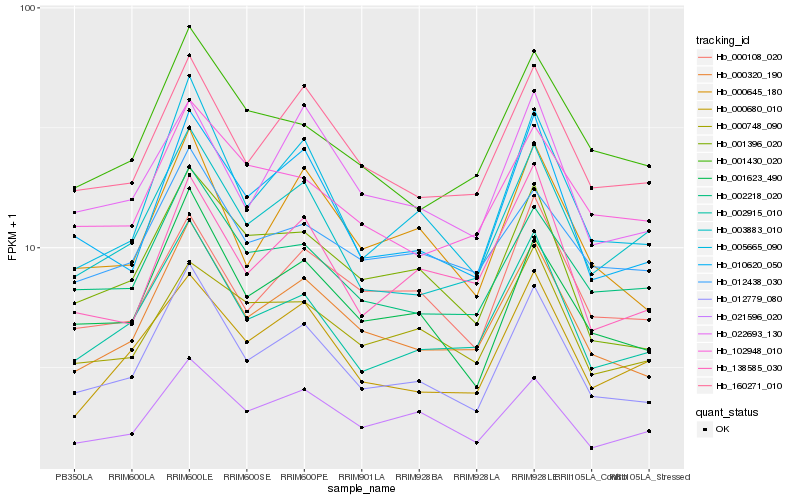
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000320_190 |
0.0 |
- |
- |
PREDICTED: aminomethyltransferase, mitochondrial [Jatropha curcas] |
| 2 |
Hb_012779_080 |
0.0546403414 |
- |
- |
PREDICTED: carbon catabolite repressor protein 4 homolog 3 isoform X3 [Jatropha curcas] |
| 3 |
Hb_160271_010 |
0.0664386221 |
- |
- |
PREDICTED: FAD synthase-like isoform X2 [Jatropha curcas] |
| 4 |
Hb_002915_010 |
0.0768823518 |
- |
- |
PREDICTED: transcriptional activator DEMETER isoform X1 [Jatropha curcas] |
| 5 |
Hb_001396_020 |
0.0810353261 |
- |
- |
glucose-6-phosphate dehydrogenase [Hevea brasiliensis] |
| 6 |
Hb_000645_180 |
0.0818969378 |
rubber biosynthesis |
Gene Name: Dihydrolipoamide dehydrogenase |
Lipoamide dehydrogenase 1 isoform 1 [Theobroma cacao] |
| 7 |
Hb_005665_090 |
0.082399226 |
- |
- |
PREDICTED: 6-phosphofructo-2-kinase/fructose-2,6-bisphosphatase isoform X2 [Jatropha curcas] |
| 8 |
Hb_010620_050 |
0.0870704377 |
- |
- |
PREDICTED: uncharacterized protein LOC105646119 [Jatropha curcas] |
| 9 |
Hb_001623_490 |
0.0880693836 |
- |
- |
PREDICTED: haloacid dehalogenase-like hydrolase domain-containing protein At2g33255 isoform X2 [Jatropha curcas] |
| 10 |
Hb_001430_020 |
0.0883180267 |
- |
- |
Actin-like ATPase superfamily protein isoform 1 [Theobroma cacao] |
| 11 |
Hb_012438_030 |
0.0915955501 |
- |
- |
PREDICTED: protein sym-1 [Jatropha curcas] |
| 12 |
Hb_002218_020 |
0.0918105983 |
- |
- |
PREDICTED: AT-hook motif nuclear-localized protein 14 [Jatropha curcas] |
| 13 |
Hb_021596_020 |
0.0921713357 |
- |
- |
hypothetical protein JCGZ_02034 [Jatropha curcas] |
| 14 |
Hb_138585_030 |
0.0943837497 |
- |
- |
PREDICTED: probable serine/threonine-protein kinase At1g54610 [Jatropha curcas] |
| 15 |
Hb_022693_130 |
0.0946400792 |
- |
- |
BRASSINOSTEROID INSENSITIVE 1-associated receptor kinase 1 precursor, putative [Ricinus communis] |
| 16 |
Hb_000680_010 |
0.0952143306 |
- |
- |
PREDICTED: putative hydrolase C777.06c [Populus euphratica] |
| 17 |
Hb_000108_020 |
0.096353654 |
- |
- |
hypothetical protein POPTR_0019s02430g [Populus trichocarpa] |
| 18 |
Hb_102948_010 |
0.0971461678 |
transcription factor |
TF Family: ARID |
PREDICTED: AT-rich interactive domain-containing protein 5 isoform X2 [Jatropha curcas] |
| 19 |
Hb_003883_010 |
0.0975294611 |
- |
- |
PREDICTED: cold-inducible RNA-binding protein [Jatropha curcas] |
| 20 |
Hb_000748_090 |
0.0978423724 |
- |
- |
glucose inhibited division protein A, putative [Ricinus communis] |