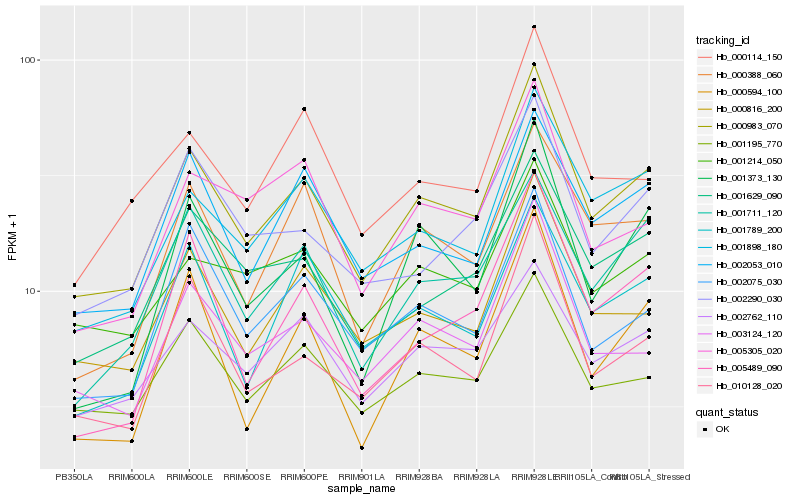
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000983_070 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_001898_180 |
0.0797493212 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_010128_020 |
0.0813561048 |
- |
- |
hypothetical protein POPTR_0001s24210g [Populus trichocarpa] |
| 4 |
Hb_000816_200 |
0.0823238564 |
- |
- |
2-deoxyglucose-6-phosphate phosphatase, putative [Ricinus communis] |
| 5 |
Hb_003124_120 |
0.0842069319 |
- |
- |
PREDICTED: fructose-1,6-bisphosphatase, cytosolic [Jatropha curcas] |
| 6 |
Hb_001629_090 |
0.0856391279 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_002053_010 |
0.0946447957 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 8 |
Hb_001214_050 |
0.0946456757 |
- |
- |
PREDICTED: bifunctional monothiol glutaredoxin-S16, chloroplastic [Jatropha curcas] |
| 9 |
Hb_001195_770 |
0.095003552 |
- |
- |
PREDICTED: probable E3 ubiquitin-protein ligase ARI2 [Jatropha curcas] |
| 10 |
Hb_001711_120 |
0.0952202502 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 11 |
Hb_002762_110 |
0.0960733266 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_002075_030 |
0.0969078512 |
- |
- |
PREDICTED: glycerol-3-phosphate acyltransferase, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000388_060 |
0.097571688 |
- |
- |
fructokinase [Manihot esculenta] |
| 14 |
Hb_002290_030 |
0.0990456511 |
- |
- |
PREDICTED: uncharacterized protein LOC105635877 isoform X1 [Jatropha curcas] |
| 15 |
Hb_005489_090 |
0.1003682315 |
- |
- |
PREDICTED: peptidyl-tRNA hydrolase ICT1, mitochondrial [Jatropha curcas] |
| 16 |
Hb_000594_100 |
0.1011557634 |
- |
- |
PREDICTED: methionine aminopeptidase 1B, chloroplastic [Jatropha curcas] |
| 17 |
Hb_001789_200 |
0.1014825417 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 18 |
Hb_001373_130 |
0.101535984 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 19 |
Hb_000114_150 |
0.1016393061 |
rubber biosynthesis |
Gene Name: Geranyl geranyl diphosphate synthase |
geranylgeranyl-diphosphate synthase [Hevea brasiliensis] |
| 20 |
Hb_005305_020 |
0.1035483052 |
- |
- |
PREDICTED: dicarboxylate transporter 1, chloroplastic [Jatropha curcas] |