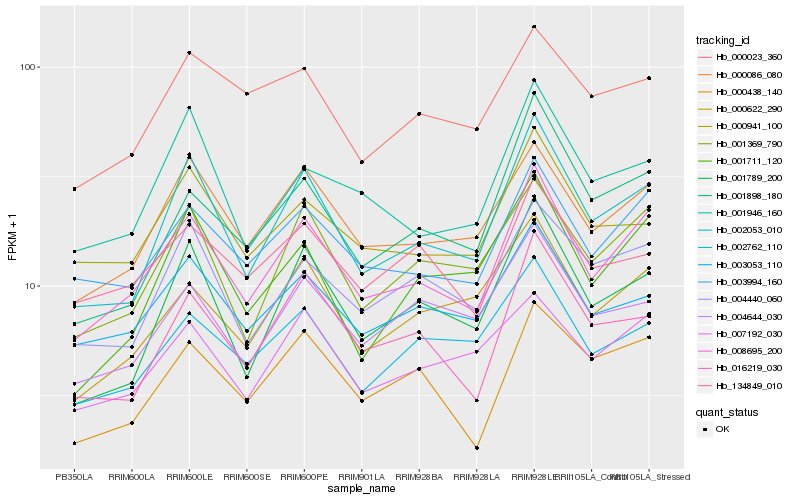
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_008695_200 |
0.0 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 2 |
Hb_003994_160 |
0.0678701207 |
- |
- |
PREDICTED: uncharacterized protein LOC105131691 isoform X5 [Populus euphratica] |
| 3 |
Hb_002053_010 |
0.0744679464 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 4 |
Hb_000086_080 |
0.0779267345 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 5 |
Hb_001369_790 |
0.087786536 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 6 |
Hb_003053_110 |
0.0955959032 |
- |
- |
PREDICTED: protease Do-like 1, chloroplastic [Jatropha curcas] |
| 7 |
Hb_007192_030 |
0.0958215733 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 8 |
Hb_000622_290 |
0.0973087842 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 9 |
Hb_000438_140 |
0.098440404 |
- |
- |
hypothetical protein CISIN_1g018088mg [Citrus sinensis] |
| 10 |
Hb_002762_110 |
0.0990243206 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 11 |
Hb_004440_060 |
0.0990950693 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 12 |
Hb_016219_030 |
0.1018155447 |
- |
- |
cytochrome P450, putative [Ricinus communis] |
| 13 |
Hb_001789_200 |
0.1019172542 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 14 |
Hb_001898_180 |
0.1039784625 |
- |
- |
PREDICTED: translation initiation factor IF-1, chloroplastic [Jatropha curcas] |
| 15 |
Hb_000941_100 |
0.1046998231 |
- |
- |
thioredoxin-like 5 mRNA family protein [Populus trichocarpa] |
| 16 |
Hb_001946_160 |
0.1051648503 |
- |
- |
putative chaperon P13.9 [Castanea sativa] |
| 17 |
Hb_004644_030 |
0.1059306948 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 18 |
Hb_134849_010 |
0.1062406403 |
- |
- |
ATP synthase subunit d, putative [Ricinus communis] |
| 19 |
Hb_001711_120 |
0.107764321 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000023_360 |
0.1091625827 |
- |
- |
phospholipid hydroperoxide glutathione peroxidase 1, chloroplastic [Jatropha curcas] |