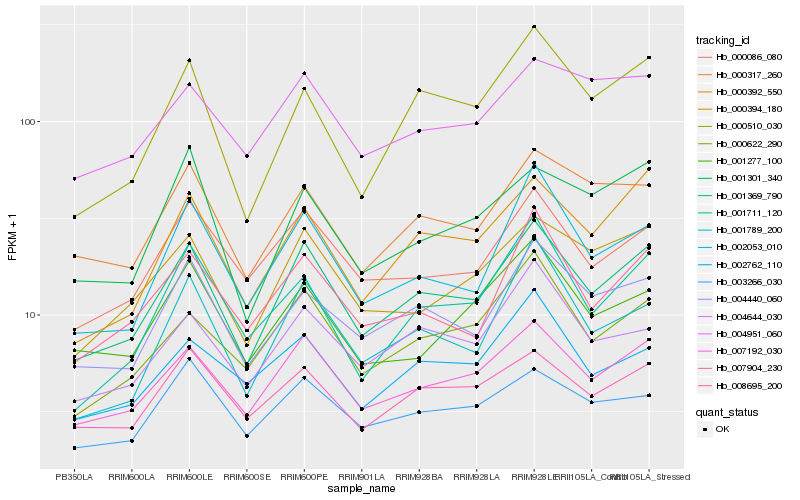
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001369_790 |
0.0 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 2 |
Hb_007192_030 |
0.0535256329 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 3 |
Hb_004440_060 |
0.0792593293 |
- |
- |
aldose 1-epimerase, putative [Ricinus communis] |
| 4 |
Hb_000392_550 |
0.0801832234 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 5 |
Hb_000086_080 |
0.0816485964 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 6 |
Hb_000394_180 |
0.0817787806 |
- |
- |
PREDICTED: uncharacterized protein LOC105636995 [Jatropha curcas] |
| 7 |
Hb_000622_290 |
0.0840802294 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |
| 8 |
Hb_002053_010 |
0.0855296672 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic-like [Jatropha curcas] |
| 9 |
Hb_007904_230 |
0.0873336671 |
- |
- |
PREDICTED: D-cysteine desulfhydrase 2, mitochondrial [Jatropha curcas] |
| 10 |
Hb_008695_200 |
0.087786536 |
- |
- |
Protein grpE, putative [Ricinus communis] |
| 11 |
Hb_000317_260 |
0.0881118307 |
- |
- |
unknown [Populus trichocarpa] |
| 12 |
Hb_001789_200 |
0.088831277 |
- |
- |
PREDICTED: sec-independent protein translocase protein TATB, chloroplastic [Jatropha curcas] |
| 13 |
Hb_000510_030 |
0.0899184207 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 14 |
Hb_004644_030 |
0.0914928607 |
- |
- |
PREDICTED: FAD synthase isoform X3 [Jatropha curcas] |
| 15 |
Hb_003266_030 |
0.0960932133 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 16 |
Hb_004951_060 |
0.0975917532 |
- |
- |
PREDICTED: uncharacterized protein LOC105634399 [Jatropha curcas] |
| 17 |
Hb_001301_340 |
0.0981865056 |
- |
- |
endoribonuclease L-PSP family protein [Populus trichocarpa] |
| 18 |
Hb_001711_120 |
0.0984762706 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 19 |
Hb_002762_110 |
0.0986334703 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 20 |
Hb_001277_100 |
0.1018568383 |
- |
- |
PREDICTED: 2-methyl-6-phytyl-1,4-hydroquinone methyltransferase, chloroplastic-like isoform X1 [Gossypium raimondii] |