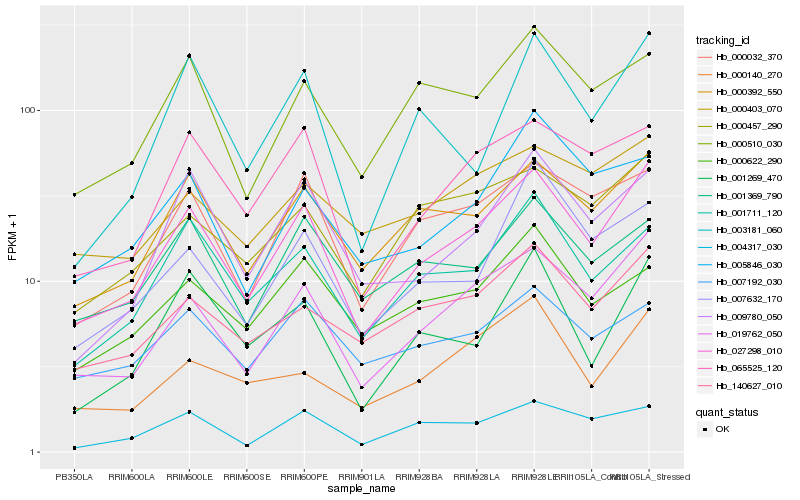
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_027298_010 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_019762_050 |
0.0664319888 |
transcription factor |
TF Family: CSD |
PREDICTED: glycine-rich protein 2-like [Jatropha curcas] |
| 3 |
Hb_000392_550 |
0.0971054511 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 6, chloroplastic isoform X1 [Jatropha curcas] |
| 4 |
Hb_000032_370 |
0.108963706 |
- |
- |
PREDICTED: nucleoside diphosphate kinase III, chloroplastic/mitochondrial-like [Citrus sinensis] |
| 5 |
Hb_065525_120 |
0.1139545411 |
- |
- |
dtdp-glucose 4-6-dehydratase, putative [Ricinus communis] |
| 6 |
Hb_140627_010 |
0.1159347426 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 7 |
Hb_009780_050 |
0.119042478 |
- |
- |
PREDICTED: glutathione S-transferase DHAR3, chloroplastic [Jatropha curcas] |
| 8 |
Hb_001711_120 |
0.1190451004 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit 4, chloroplastic [Jatropha curcas] |
| 9 |
Hb_000457_290 |
0.1214259907 |
- |
- |
ATP-dependent Clp protease proteolytic subunit, putative [Ricinus communis] |
| 10 |
Hb_004317_030 |
0.1214897265 |
- |
- |
PREDICTED: protein CHROMOSOME TRANSMISSION FIDELITY 7 [Jatropha curcas] |
| 11 |
Hb_000510_030 |
0.1259231979 |
- |
- |
Winged-helix DNA-binding transcription factor family protein [Theobroma cacao] |
| 12 |
Hb_001269_470 |
0.1289567505 |
- |
- |
PREDICTED: uncharacterized protein LOC105630313 [Jatropha curcas] |
| 13 |
Hb_001369_790 |
0.1322305589 |
- |
- |
PREDICTED: clp protease-related protein At4g12060, chloroplastic [Jatropha curcas] |
| 14 |
Hb_003181_060 |
0.1343303961 |
- |
- |
PREDICTED: triosephosphate isomerase, cytosolic [Jatropha curcas] |
| 15 |
Hb_000403_070 |
0.1359443501 |
- |
- |
PREDICTED: iron-sulfur assembly protein IscA, chloroplastic isoform X1 [Jatropha curcas] |
| 16 |
Hb_000140_270 |
0.1361774642 |
- |
- |
S-adenosyl-L-methionine-dependent methyltransferases superfamily protein [Theobroma cacao] |
| 17 |
Hb_007632_170 |
0.1378851332 |
- |
- |
hypothetical protein JCGZ_01028 [Jatropha curcas] |
| 18 |
Hb_007192_030 |
0.1394334697 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 19 |
Hb_005846_030 |
0.1401171542 |
- |
- |
PREDICTED: 50S ribosomal protein L29, chloroplastic [Jatropha curcas] |
| 20 |
Hb_000622_290 |
0.141901972 |
- |
- |
PREDICTED: F-box/kelch-repeat protein At3g61590 [Jatropha curcas] |