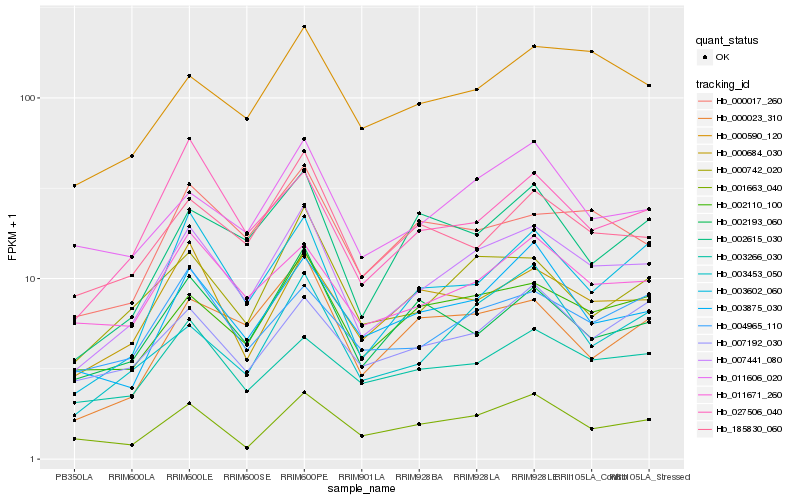
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_007441_080 |
0.0 |
- |
- |
PREDICTED: ELMO domain-containing protein C-like [Jatropha curcas] |
| 2 |
Hb_002110_100 |
0.0909410236 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 3 |
Hb_000742_020 |
0.1019765015 |
- |
- |
ARF GTPase activator, putative [Ricinus communis] |
| 4 |
Hb_000684_030 |
0.1053050607 |
- |
- |
syntaxin, putative [Ricinus communis] |
| 5 |
Hb_003602_060 |
0.1060107625 |
- |
- |
PREDICTED: phosphoglycerate mutase-like protein 1 [Jatropha curcas] |
| 6 |
Hb_000590_120 |
0.1084852173 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 7 |
Hb_003875_030 |
0.1100342345 |
- |
- |
PREDICTED: probable plastidic glucose transporter 1 [Jatropha curcas] |
| 8 |
Hb_011671_260 |
0.1104386696 |
- |
- |
PREDICTED: putative lipase ROG1 isoform X1 [Jatropha curcas] |
| 9 |
Hb_185830_060 |
0.1115775674 |
- |
- |
PREDICTED: ras-related protein RABB1c [Fragaria vesca subsp. vesca] |
| 10 |
Hb_000017_260 |
0.1116862145 |
- |
- |
hypothetical protein POPTR_0003s13070g [Populus trichocarpa] |
| 11 |
Hb_004965_110 |
0.1129041971 |
- |
- |
PREDICTED: folylpolyglutamate synthase [Jatropha curcas] |
| 12 |
Hb_027506_040 |
0.113804114 |
rubber biosynthesis |
Gene Name: Pyruvate dehydrogenase |
PREDICTED: pyruvate dehydrogenase E1 component subunit beta-3, chloroplastic [Jatropha curcas] |
| 13 |
Hb_011606_020 |
0.1145709036 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 14 |
Hb_001663_040 |
0.1172472468 |
- |
- |
hypothetical protein B456_007G162000 [Gossypium raimondii] |
| 15 |
Hb_003266_030 |
0.1180618996 |
- |
- |
hypothetical protein POPTR_0002s05550g [Populus trichocarpa] |
| 16 |
Hb_002615_030 |
0.1190131904 |
- |
- |
PREDICTED: E3 ubiquitin-protein ligase RHF1A-like [Populus euphratica] |
| 17 |
Hb_003453_050 |
0.1193622263 |
- |
- |
PREDICTED: calcium-binding mitochondrial carrier protein SCaMC-3-like [Jatropha curcas] |
| 18 |
Hb_002193_060 |
0.1194480922 |
- |
- |
PREDICTED: uncharacterized protein C4orf29 homolog [Pyrus x bretschneideri] |
| 19 |
Hb_007192_030 |
0.1203510019 |
- |
- |
PREDICTED: manganese-dependent ADP-ribose/CDP-alcohol diphosphatase [Jatropha curcas] |
| 20 |
Hb_000023_310 |
0.1203838979 |
- |
- |
PREDICTED: probable folate-biopterin transporter 2 [Populus euphratica] |