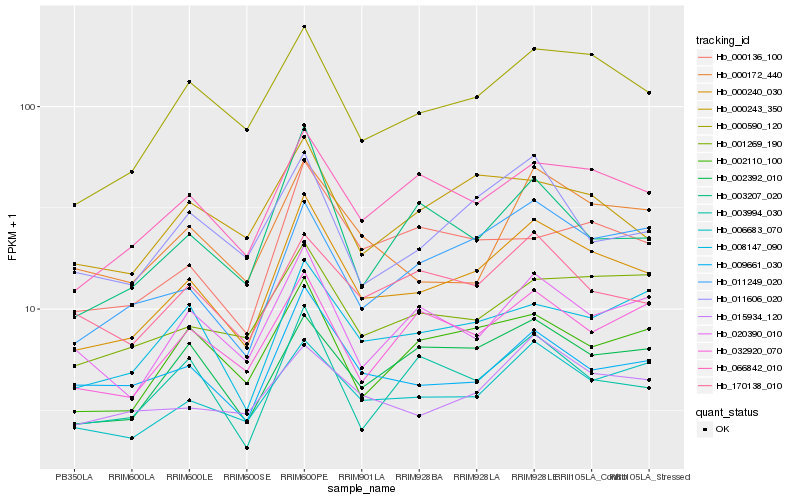
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_000240_030 |
0.0 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 2 |
Hb_000590_120 |
0.0803966979 |
- |
- |
dehydroascorbate reductase, putative [Ricinus communis] |
| 3 |
Hb_066842_010 |
0.0887067 |
- |
- |
PREDICTED: probable 6-phosphogluconolactonase 4, chloroplastic [Jatropha curcas] |
| 4 |
Hb_006683_070 |
0.0914989448 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 5 |
Hb_011249_020 |
0.0978576753 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 6 |
Hb_002392_010 |
0.1004365512 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 7 |
Hb_002110_100 |
0.1045898494 |
- |
- |
PREDICTED: pyridoxine/pyridoxamine 5'-phosphate oxidase 1, chloroplastic [Jatropha curcas] |
| 8 |
Hb_009661_030 |
0.1048988461 |
- |
- |
PREDICTED: SPX and EXS domain-containing protein 1-like [Jatropha curcas] |
| 9 |
Hb_001269_190 |
0.1060974837 |
- |
- |
PREDICTED: Golgi SNAP receptor complex member 1-1 [Jatropha curcas] |
| 10 |
Hb_015934_120 |
0.106393055 |
- |
- |
PREDICTED: callose synthase 7 [Vitis vinifera] |
| 11 |
Hb_008147_090 |
0.1067612093 |
- |
- |
conserved hypothetical protein [Ricinus communis] |
| 12 |
Hb_020390_010 |
0.1091534338 |
- |
- |
PREDICTED: expansin-A13 [Jatropha curcas] |
| 13 |
Hb_000136_100 |
0.1103263907 |
- |
- |
PREDICTED: syntaxin-61 [Jatropha curcas] |
| 14 |
Hb_003994_030 |
0.1107202587 |
- |
- |
PREDICTED: organic cation/carnitine transporter 7 [Jatropha curcas] |
| 15 |
Hb_000172_440 |
0.1108964735 |
- |
- |
PREDICTED: NADPH-dependent 1-acyldihydroxyacetone phosphate reductase [Jatropha curcas] |
| 16 |
Hb_011606_020 |
0.111227565 |
- |
- |
hypothetical protein CISIN_1g023687mg [Citrus sinensis] |
| 17 |
Hb_000243_350 |
0.1129803491 |
- |
- |
PREDICTED: V-type proton ATPase 16 kDa proteolipid subunit [Tarenaya hassleriana] |
| 18 |
Hb_003207_020 |
0.1129879705 |
- |
- |
PREDICTED: bifunctional dTDP-4-dehydrorhamnose 3,5-epimerase/dTDP-4-dehydrorhamnose reductase [Jatropha curcas] |
| 19 |
Hb_032920_070 |
0.1135850605 |
- |
- |
PREDICTED: zinc finger protein-like 1 homolog [Jatropha curcas] |
| 20 |
Hb_170138_010 |
0.1137128912 |
- |
- |
conserved hypothetical protein [Ricinus communis] |