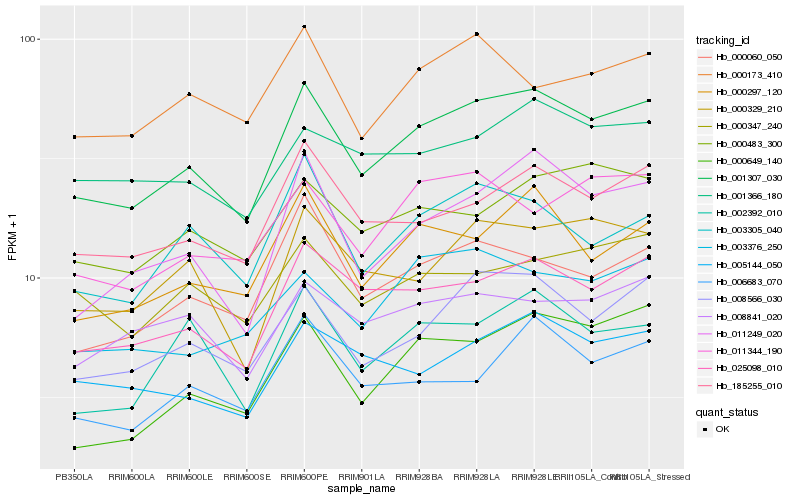
| Rank |
Gene |
Score (JSD) |
Function |
Description |
NCBI(nr) information |
| 1 |
Hb_001307_030 |
0.0 |
- |
- |
PREDICTED: succinyl-CoA ligase [ADP-forming] subunit beta, mitochondrial [Jatropha curcas] |
| 2 |
Hb_025098_010 |
0.0657881281 |
- |
- |
PREDICTED: protoheme IX farnesyltransferase, mitochondrial isoform X2 [Jatropha curcas] |
| 3 |
Hb_008566_030 |
0.0662814312 |
- |
- |
hypothetical protein POPTR_0013s02780g [Populus trichocarpa] |
| 4 |
Hb_185255_010 |
0.0742149128 |
- |
- |
PREDICTED: DEAD-box ATP-dependent RNA helicase 11-like [Jatropha curcas] |
| 5 |
Hb_011249_020 |
0.0767313463 |
- |
- |
PREDICTED: putative clathrin assembly protein At5g35200 [Jatropha curcas] |
| 6 |
Hb_000297_120 |
0.0772785601 |
- |
- |
PREDICTED: sorting nexin 1 [Jatropha curcas] |
| 7 |
Hb_000329_210 |
0.0831767969 |
- |
- |
Golgi snare 12 isoform 1 [Theobroma cacao] |
| 8 |
Hb_005144_050 |
0.0849042021 |
- |
- |
PREDICTED: S-adenosylmethionine synthase 1 isoform X2 [Vitis vinifera] |
| 9 |
Hb_000060_050 |
0.0857177174 |
- |
- |
PREDICTED: uncharacterized protein LOC105649718 [Jatropha curcas] |
| 10 |
Hb_003305_040 |
0.087873057 |
- |
- |
AP47/50p mRNA family protein [Populus trichocarpa] |
| 11 |
Hb_002392_010 |
0.0889720619 |
- |
- |
PREDICTED: apurinic endonuclease-redox protein isoform X4 [Jatropha curcas] |
| 12 |
Hb_000483_300 |
0.0899602683 |
- |
- |
PREDICTED: ATP-dependent Clp protease proteolytic subunit-related protein 4, chloroplastic [Populus euphratica] |
| 13 |
Hb_001366_180 |
0.090371952 |
- |
- |
PREDICTED: translocase of chloroplast 33, chloroplastic-like [Jatropha curcas] |
| 14 |
Hb_003376_250 |
0.0913189097 |
- |
- |
Transducin/WD40 repeat-like superfamily protein [Theobroma cacao] |
| 15 |
Hb_008841_020 |
0.0913785212 |
transcription factor |
TF Family: PHD |
PREDICTED: uncharacterized protein LOC105631075 isoform X1 [Jatropha curcas] |
| 16 |
Hb_006683_070 |
0.0916557634 |
- |
- |
PREDICTED: uncharacterized protein LOC105631443 [Jatropha curcas] |
| 17 |
Hb_011344_190 |
0.0925579325 |
- |
- |
PREDICTED: maspardin [Jatropha curcas] |
| 18 |
Hb_000173_410 |
0.0944392095 |
- |
- |
PREDICTED: BI1-like protein [Jatropha curcas] |
| 19 |
Hb_000347_240 |
0.0945605647 |
- |
- |
PREDICTED: proteasome subunit alpha type-6 [Pyrus x bretschneideri] |
| 20 |
Hb_000649_140 |
0.0952159723 |
transcription factor |
TF Family: SET |
PREDICTED: histone-lysine N-methyltransferase setd3 isoform X1 [Jatropha curcas] |